
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Monica Neagu | + 1739 word(s) | 1739 | 2021-12-20 09:53:27 | | | |
| 2 | Lindsay Dong | Meta information modification | 1739 | 2021-12-20 10:57:57 | | |
Video Upload Options
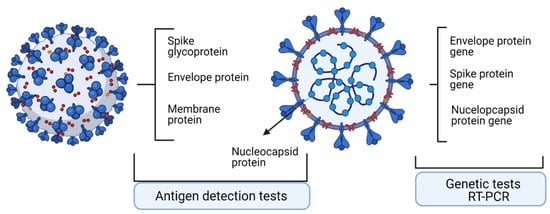
In diagnosing SARS-CoV-2 infection, the most widely used test is the molecular testing. Real-time reverse transcription polymerase chain reaction (RT-PCR) is the most well-known and extensively used molecular analysis. The test relies on nucleic acid amplification and detects unique sequences of SARS-CoV-2. The other type of test, the antigen tests, can detect the presence of SARS-CoV-2 without amplifying viral components, but these tests are less sensitive than the molecular ones. Commonly, any negative antigen test is confirmed with a molecular test so that the patient can be declared negative for COVID-19. Both molecular and antigen tests would detect patients in the acute phase of infection.
1. Introduction
Laboratory diagnosis in COVID-19 is influential in combating the spreading of SARS-CoV-2 infection. Moreover, laboratory tests dictate the clinical decisions regarding the infected patient. These tests comprise the ones that detect the viral genome and testes that detect the viral proteome. Upon molecular and antigen tests, patients were classified as positive or negative for the presence of SARS-CoV-2. Nevertheless, all tests have two seminal characteristics/parameters, namely, percent positive agreement (PPA), describing the actual sensitivity of the test, and percent negative agreement (PNA), describing the specificity of the test [1].
In diagnosing SARS-CoV-2 infection, the most widely used test is the molecular testing. Real-time reverse transcription polymerase chain reaction (RT-PCR) is the most well-known and extensively used molecular analysis. The test relies on nucleic acid amplification and detects unique sequences of SARS-CoV-2 [2]. The other type of test, the antigen tests, can detect the presence of SARS-CoV-2 without amplifying viral components, but these tests are less sensitive than the molecular ones. Commonly, any negative antigen test is confirmed with a molecular test so that the patient can be declared negative for COVID-19. Both molecular and antigen tests would detect patients in the acute phase of infection [3][4].
2. Technologies to Assess Specific Antigens
2.1. Quantitative Real-Time Reverse Transcriptase-PCR
2.2. Nonconventional Tests—Droplet-Digital PCR
2.3. Antigen Detection Tests
| Test Type | Advantages | Disadvantages | Test Sensitivity % | Test Specificity % |
|---|---|---|---|---|
| Test for viral genome | Accurate tests, identifies mutations in the virus, it tracks disease spread. | Does not detect viral load, does not detect dynamics of infection or the history of prior infection. | 86.1% | 95.8% |
| Test for viral antigen | Detects proteins on the viral particle surface. | Less sensitive than molecular tests and often a molecular test need to confirm the positive result. | 61.7% | 98.2% |
| Faster than molecular tests, less expensive, applicable to large number of samples. |
References
- Brooks, Z.C.; Saswati, D. COVID-19 Testing Impact of Prevalence, Sensitivity, and Specificity on Patient Risk and Cost. Am. J. Clin. Pathol. 2020, 154, 575–584.
- Lippi, G.; Sanchis-Gomar, F.; Henry, B.M. COVID-19: Unravelling the clinical progression of nature’s virtually perfect biological weapon. Ann. Transl. Med. 2020, 8, 693.
- Food and Drug Administration. EUA Authorized Serology Test Performance. Available online: https://www.fda.gov/medical-devices/mergency-situations-medical-devices/eua-authorized-serologytest-performance (accessed on 7 September 2021).
- UK Medicines & Healthcare Products Regulatory Agency. Target Product Profile: Antibody Tests to Help Determine if People Have Recent Infection to SARSCoV-2. Available online: https://www.gov.uk/government/publications/how-testsand-testing-kits-for-coronavirus-covid-19-work/target-product-profile-antibody-tests-to-help-determine-ifpeople-have-recent-infection-to-sars-cov-2-version-2 (accessed on 15 August 2021).
- Stites, E.C.; Wilen, C.B. The Interpretation of SARS-CoV-2 Diagnostic Tests. Med NY 2020, 1, 78–89.
- CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. 2020. Available online: https://www.fda.gov/media/134922/download (accessed on 20 December 2020).
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Marie Luisa Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2001035.
- Weissleder, R.; Lee, H.; Ko, J.; Pittet, M.J. COVID-19 diagnostics in context. Sci. Transl. Med. 2020, 12, eabc1931.
- Leong, H.N.; Chan, K.P.; Khan, A.S.; Oon, L.; Su Yun Se-Thoe, S.Y.S.-T.; Bai, X.L.B.; Yeo, D.; Leo, Y.S.; Ang, B.; Ksiazek, T.G.; et al. Virus-specific RNA and antibody from convalescent-phase SARS patients discharged from hospital. Emerg. Infect. Dis. 2004, 10, 1745–1750.
- He, X.; Lau, E.H.Y.; Wu, P.; Deng, X.; Wang, J.; Hao, X.; Lau, Y.C.; Wong, J.Y.; Guan, Y.; Tan, X.; et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020, 26, 672–675.
- Huang, A.T.; Garcia-Carreras, B.; Hitchings, M.D.T.; Yang, B.; Katzelnick, L.C.; Rattigan, S.M.; Borgert, B.A.; Moreno, C.A.; Solomon, B.D.; Rodriguez-Barraquer, I.; et al. A systematic review of antibody mediated immunity to coronaviruses: Antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. Nat. Commun. 2020, 11, 4704–4770.
- Kaye, M. SARS-associated coronavirus replication in cell lines. Emerg. Infect. Dis. 2006, 12, 128–133.
- Yu, F.; Yan, L.; Wang, N.; Yang, S.; Wang, L.; Tang, Y.; Gao, G.; Wang, S.; Ma, C.; Xie, R.; et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin. Infect. Dis. 2020, 71, 793–798.
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020, 323, 1843–1844.
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469.
- Mao, X.; Liu, C.; Tong, H.; Chen, Y.; Liu, K. Principles of digital PCR and its applications in current obstetrical and gynecological diseases. Am. J. Transl. Res. 2019, 11, 7209–7222.
- Manoj, P. Droplet digital PCR technology promises new applications and research areas. Mitochondrial DNA 2016, 27, 742–746.
- Tong, Y.; Shen, S.; Jiang, H.; Chen, A.Z. Application of Digital PCR in Detecting Human Diseases Associated Gene Mutation. Cell. Physiol. Biochem. 2017, 43, 1718–1730.
- McEvoy, A.C.; Wood, B.A.; Ardakani, N.M.; Pereira, M.R.; Pearce, R.; Cowell, L.; Robinson, C.; Grieu-Iacopetta, F.; Spicer, A.J.; Amanuel, B.; et al. Droplet Digital PCR for Mutation Detection in Formalin-Fixed, Paraffin-Embedded Melanoma Tissues: A Comparison with Sanger Sequencing and Pyrosequencing. J. Mol. Diagn. 2018, 20, 240–252.
- Dobre, E.-G.; Neagu, M. Multi-Omics-Driven Biomarkers for Precision Medicine in Cutaneous Melanoma. In Proceedings of the 1st International Electronic Conference on Biomedicine, Rensselae, NY, USA, 1–26 March 2021.
- Park, C.; Lee, J.; Hassan, Z.U.; Ku, K.B.; Kim, S.J.; Kim, H.G.; Park, E.C.; Park, G.S.; Park, D.; Baek, S.H.; et al. Comparison of Digital PCR and Quantitative PCR with Various SARS-CoV-2 Primer-Probe Sets. J. Microbiol. Biotechnol. 2021, 31, 358–367.
- De Kock, R.; Baselmans, M.; Scharnhorst, V.; Deiman, B. Sensitive detection and quantification of SARS-CoV-2 by multiplex droplet digital RT-PCR. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 807–813.
- Deiana, M.; Mori, A.; Piubelli, C.; Scarso, S.; Favarato, M.; Elena Pomari, E. Assessment of the direct quantitation of SARS-CoV-2 by droplet digital PCR. Sci. Rep. 2020, 10, 18764–18771.
- Tedim, A.P.; Almansa, R.; Domínguez-Gil, M.; González-Rivera, M.; Micheloud, D.; Ryan, P.; Méndez, R.; Blanca-López, N.; Pérez-García, F.; Bustamante, E.; et al. Comparison of real-time and droplet digital PCR to detect and quantify SARS-CoV-2 RNA in plasma. Eur. J. Clin. Investig. 2021, 51, e13501.
- Moore, B.J.B.; June, C.H. Cytokine release syndrome in severe COVID-19. Science 2020, 368, 473–474.




