
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Piotr Kozlowski | + 2652 word(s) | 2652 | 2020-07-24 11:46:14 | | | |
| 2 | Piotr Kozlowski | + 6 word(s) | 2658 | 2020-07-28 12:05:45 | | | | |
| 3 | Rita Xu | -1019 word(s) | 1639 | 2020-07-30 10:40:00 | | |
Video Upload Options
A catalog of BARD1 germline mutations/pathogenic variants (PVs) identified in large cumulative cohorts of ~48,700 breast cancer (BC) and ~20,800 ovarian cancer (OC) cases prepared based on 123 studies published over 20 years of BARD1 gene screening. By comparing the frequency of BARD1 PVs in the cases and ~134,100 noncancer controls from the gnomAD database, the effect of the BARD1 PVs on BC and OC risks is estimated.
1. Definition
Over the last two decades, numerous BARD1 mutations/pathogenic variants (PVs) have been found in patients with breast cancer (BC) and ovarian cancer (OC). However, their role in BC and OC susceptibility remains controversial, and strong evidence-based guidelines for carriers are not yet available.
2. Introduction
Inherited genetic factors are responsible for a substantial portion of breast cancer (BC) and ovarian cancer (OC) cases. Multigene panel testing has revealed that germline loss-of-function mutations/pathogenic variants (PVs) in various cancer-associated genes are identified in more than 10% and 20% of patients with BC and OC, respectively [1][2][3]. Attempts have been made to define the genetic/familial risk of BC/OC associated with these genes, and subsequently, management recommendations for carriers of PVs in some of the high-penetrance genes have been established [4][5]. Despite this, several genes in which PVs confer low- or moderate-penetrance effects still require more evidence and more convincing assessments of BC/OC risk to utilize them in recommendations for carriers. A group of genes with insufficient and/or conflicting data includes the BRCA1-associated RING domain 1 (BARD1) gene.
The BARD1 gene encodes a protein of 777 amino acids with several functional domains, including an N-terminal RING-finger domain, three ankyrin repeats (ANK), and two C-terminal BRCT domains. The protein shows structural homology with BRCA1 within the BRCT and RING-finger domains, and through the latter domains, the two proteins form a stable heterodimer. The BARD1-BRCA1 heterodimer has E3 ubiquitin ligase activity and acts in multiple cellular processes essential for maintaining genomic stability, including DNA double-strand break repair through homologous recombination [6][7][8]. In addition to the well-established role of the BARD1-BRCA1 heterodimer, the BRCA1-independent function of BARD1 as a tumor suppressor has also been postulated, for example, in mediating p53-dependent apoptosis [9]. On the other hand, several BARD1 isoforms resulting from alternative splicing that lack RING-finger and/or ANK domains were upregulated in different cancers and are suggested to have an oncogenic effect by antagonizing the function of full-length BARD1 [10][11][12][13].
Because of the abovementioned BARD1 functions, many mutation screening studies have been carried out to explore the role of BARD1 in BC and OC susceptibility. As a result, hundreds of PVs were detected, mostly through the use of high-throughput next-generation sequencing (NGS; panel sequencing) in recent years. However, the PVs are scattered among many articles, which prevents the drawing of more comprehensive conclusions about their distribution over the gene, identification of recurrent PVs, and estimation of a more precise effect on cancer risk. In addition, despite multiple examples of BARD1 PVs in BC/OC patients, attempts to link PVs with BC/OC risk are often inconclusive and are usually not supported by statistically convincing pieces of evidence. However, these types of studies are justified due to reduced mutation penetrance and various interactions between the gene and other genetic, personal, and environmental factors. The lack of proper functional characterization of PVs and the genetic heterogeneity in different populations may be additional factors of insufficient and/or conflicting data. In addition, due to the low frequency of BARD1 PVs (<1%), the sample sizes of the studies are still too modest to achieve sufficient statistical power, although the sizes are constantly increasing. Additionally, as the cost of whole gene testing is still relatively high and association case-control studies relying on the use of geographically matched controls are still very rare [14][15][16][17]. Therefore, controls from publicly available databases are frequently used to assess the risk, allowing to increase the study size and to improve statistical power [3][18][19][20]. Cosegregation studies can provide additional evidence for or against the association, although they may be challenging for low or moderate penetrance PVs, especially because large families with multiple BC/OC cases are becoming less available.
To overcome the limitations mentioned above, the BARD1 PVs identified in a total of ~48,700 BC and ~20,800 OC patients (from 123 studies examining the whole coding sequence of BARD1) have been cataloged. Using the collected data, the overall BC and OC risk associated with PVs in BARD1 have been determined with high confidence.
3. BARD1 Mutational Spectrum
In total, 144 BARD1 PVs (69 distinct PVs), were identified in either BC or OC cases, constituting 120 PVs (60 distinct PVs) in 48,717 BC cases (0.25%) and 24 PVs (17 distinct PVs) in 20,799 OC cases (0.12%). Out of 69 distinct PVs in the BC and OC cohorts, 21 were present in population controls [21]. The distributions of mutation types were similar in BC and OC, with nonsense, frameshift, splicing, and pathogenic missense variants accounting for 54%, 33%, 11%, and ~2% in BC and 54%, 38%, 8%, and 0% in OC, respectively. For comparison, in population controls, nonsense, frameshift, and splicing variants accounted for 47%, 43%, and 10%, respectively. The distribution of all the PVs in BARD1 detected in the BC and OC cohorts, as well as in population controls, is presented in Figure 1.
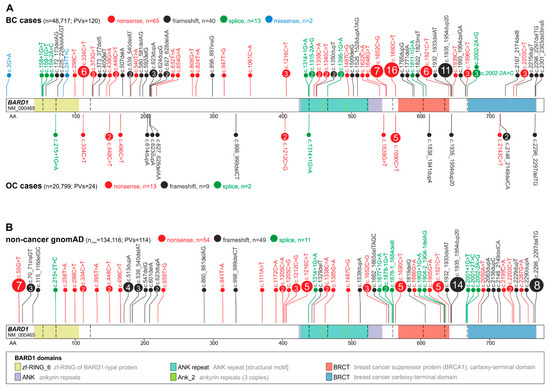
Figure 1. Maps of deleterious PVs in BARD1. PVs are shown alongside the BARD1 coding sequence with the indicated exon structure and the protein functional domains. The size of a PV symbol (circle) is proportional to the number of PVs, and color indicates the type of PV. (A) PVs detected in BC (above) and OC (below) cases. (B) PVs reported in noncancer gnomAD controls. The total number of detected PVs and the total number of cases and controls tested for the variants are indicated in parentheses; note that the total number of tested subjects differs substantially between groups.
Although BARD1 PVs in BC and OC patients are distributed over the entire coding sequence, there are two regions of increased density of PVs, i.e., from exons 2 to ~230AA in exon 4, overlapping the RING-finger domain and from exon 5 to exon 10, overlapping the ANK repeat and BRCT I domains.
In the BC cohort, 10 of the PVs were identified in three or more cases (defined here as recurrent), with the highest occurrence of c.1690C>T (Q564*), c.1935_1954dup20 (E652Vfs*69), c.1652C>G (S551*), c.334C>T (R112*), and c.1921C>T (R641*) reported in 16, 11, 7, 6, and 6 cases, respectively. R112* and R641* recurrent PVs were identified in both Caucasian and Asian populations, while the remaining PVs, including the most frequent, Q564* and E652Vfs*69, were identified only in the Caucasian population. None of the recurrent PVs were specific to the Asian population. Interestingly, 10 out of 16 Q564* PVs were reported by studies analyzing mainly patients with a family history of BC/OC. Only one recurrent PV, i.e., Q564*, was reported in the OC cohort (five patients). In population controls, an additional recurrent PV was observed (not identified in BC/OC cohorts), i.e., c.55G>T (E19*). However, the PV was characteristic only for the Latino population, which represents a small proportion of the entire population of BC/OC cohorts.
4. Association of BARD1 Pathogenic Variants with Breast Cancer
The prevalence of BARD1 PVs was higher in BC patients (0.25%) than in population controls (0.09%), with a cumulative OR = 2.90 (95% CIs:2.25–3.75; p < 0.0001), classifying BARD1 as a moderate BC risk gene . The risk was slightly higher for familial BC patients (OR = 3.67; 95% CI:2.52–5.34; p < 0.0001), evaluated based on data from studies that mainly analyzed cases with a BC/OC family history. To check whether the mixed population affected the results, similar association analyses were performed separately for Caucasian and Asian populations. As shown in Figure 2, BC risk estimates for Caucasians and Asians do not differ substantially, and both are only slightly lower than the risk estimates for mixed populations (OR = 2.73; 95% CIs:1.94–3.86; p < 0.0001 for Caucasians and OR = 2.50; 95% CIs:1.43–4.35; p = 0.0012 for Asians). The calculation was repeated excluding the three large studies from the analysis, which reported results of multigene testing of samples tested in diagnostic companies [22][23][24], as some small fractions of these samples may represent unrecognized duplicates of patients analyzed in other studies. Excluding these studies did not change the risk estimates substantially (OR = 3.14; 95% CIs:2.41–4.09; p < 0.0001). Moreover, to estimate the potential bias associated with the use of older technologies, which may be less sensitive when detecting some variants, the analysis was repeated including only studies reporting variants detected using NGS; again, the risk estimates showed no substantial difference (OR = 2.83; 95% CIs:2.18–3.67; p < 0.0001).
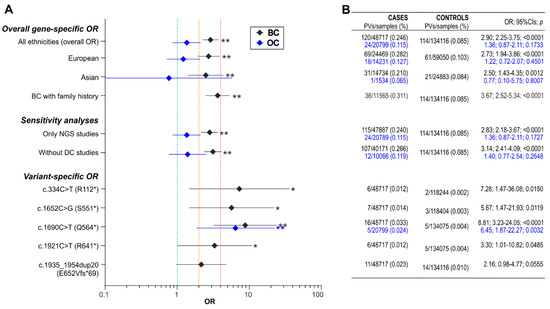
Figure 2. Summary of the BC and OC risk (OR) associated with BARD1 PVs. (A) The graph showing gene-specific and variant-specific ORs. The gene-specific OR is provided for all ethnicities combined and separately for European and Asian populations. “Only NGS studies” and “Without DC studies” demonstrate the results of repeated association analyses without studies applying older technologies and without studies publishing results of multigene testing in samples submitted to diagnostic companies, respectively. Diamonds and horizontal lines indicate the OR values and 95% CIs, respectively. Black and blue symbols represent BC and OC, respectively. Green, orange, and red vertical lines highlight an OR of 1 (no risk), an OR of 2 (the threshold for moderate risk), and an OR of 4 (the threshold for high risk), respectively. * or ** next to the OR symbol indicates a p-value < 0.05 or < 0.01, respectively. (B) The table showing the exact numbers and percentages (in the bracket) of detected PVs in either BC (black fonts) or OC (blue fonts) cases and in controls, as well as ORs with 95% CIs and p-values (the values correspond to the particular OR symbols shown in the graph on the left).
The prevalence of BARD1 PVs was only slightly higher in OC patients (0.12%) than in population controls (0.09%), with a slight non-significantly increased cumulative OR = 1.36 (95% CIs:0.87–2.11; p = 0.1733). Therefore, the association was insufficient to classify BARD1 as a low-risk gene for OC (see Figure 2 for detailed analysis).
References
- Suszynska, M.; Klonowska, K.; Jasinska, A.J.; Kozlowski, P. Large-scale meta-analysis of mutations identified in panels of breast/ovarian cancer-related genes—Providing evidence of cancer predisposition genes. Gynecol. Oncol. 2019, 153, 452–462.
- Walsh, T.; Casadei, S.; Lee, M.K.; Pennil, C.C.; Nord, A.S.; Thornton, A.M.; Roeb, W.; Agnew, K.J.; Stray, S.M.; Wickramanayake, A.; et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 18032–18037.
- Couch, F.J.; Shimelis, H.; Hu, C.; Hart, S.N.; Polley, E.C.; Na, J.; Hallberg, E.; Moore, R.; Thomas, A.; Lilyquist, J.; et al. Associations Between Cancer Predisposition Testing Panel Genes and Breast Cancer. JAMA Oncol. 2017, 3, 1190–1196.
- Daly, M.B.; Pilarski, R.; Yurgelun, M.B.; Berry, M.P.; Buys, S.S.; Dickson, P.; Domchek, S.M.; Elkhanany, A.; Friedman, S.; Garber, J.E.; et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 1.2020. J. Natl. Compr. Canc. Netw. 2020, 18, 380–391.
- Daly, M.B.; Pilarski, R.; Berry, M.; Buys, S.S.; Farmer, M.; Friedman, S.; Garber, J.E.; Kauff, N.D.; Khan, S.; Klein, C.; et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast and Ovarian, Version 2.2017. J. Natl. Compr. Canc. Netw. 2017, 15, 9–20.
- Hashizume, R.; Fukuda, M.; Maeda, I.; Nishikawa, H.; Oyake, D.; Yabuki, Y.; Ogata, H.; Ohta, T. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 2001, 276, 14537–14540.
- Irminger-Finger, I.; Ratajska, M.; Pilyugin, M. New concepts on BARD1: Regulator of BRCA pathways and beyond. Int. J. Biochem. Cell Biol. 2016, 72, 1–17.
- Tarsounas, M.; Sung, P. The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication. Nat. Rev. Mol. Cell Biol. 2020, 21, 284–299.
- Irminger-Finger, I.; Leung, W.C.; Li, J.; Dubois-Dauphin, M.; Harb, J.; Feki, A.; Jefford, C.E.; Soriano, J.V.; Jaconi, M.; Montesano, R.; et al. Identification of BARD1 as mediator between proapoptotic stress and p53-dependent apoptosis. Mol. Cell 2001, 8, 1255–1266.
- Cimmino, F.; Formicola, D.; Capasso, M. Dualistic Role of BARD1 in Cancer. Genes (Basel) 2017, 8, 375.
- Li, L.; Ryser, S.; Dizin, E.; Pils, D.; Krainer, M.; Jefford, C.E.; Bertoni, F.; Zeillinger, R.; Irminger-Finger, I. Oncogenic BARD1 isoforms expressed in gynecological cancers. Cancer Res. 2007, 67, 11876–11885.
- Zhang, Y.Q.; Bianco, A.; Malkinson, A.M.; Leoni, V.P.; Frau, G.; De Rosa, N.; Andre, P.A.; Versace, R.; Boulvain, M.; Laurent, G.J.; et al. BARD1: An independent predictor of survival in non-small cell lung cancer. Int. J. Cancer 2012, 131, 83–94.
- Bosse, K.R.; Diskin, S.J.; Cole, K.A.; Wood, A.C.; Schnepp, R.W.; Norris, G.; Nguyen le, B.; Jagannathan, J.; Laquaglia, M.; Winter, C.; et al. Common variation at BARD1 results in the expression of an oncogenic isoform that influences neuroblastoma susceptibility and oncogenicity. Cancer Res. 2012, 72, 2068–2078.
- Suszynska, M.; Kluzniak, W.; Wokolorczyk, D.; Jakubowska, A.; Huzarski, T.; Gronwald, J.; Debniak, T.; Szwiec, M.; Ratajska, M.; Klonowska, K.; et al. BARD1 is A Low/Moderate Breast Cancer Risk Gene: Evidence Based on An Association Study of the Central European p.Q564X Recurrent Mutation. Cancers (Basel) 2019, 11, 740.
- Weber-Lassalle, N.; Borde, J.; Weber-Lassalle, K.; Horvath, J.; Niederacher, D.; Arnold, N.; Kaulfuss, S.; Ernst, C.; Paul, V.G.; Honisch, E.; et al. Germline loss-of-function variants in the BARD1 gene are associated with early-onset familial breast cancer but not ovarian cancer. Breast Cancer Res. 2019, 21, 55.
- Ramus, S.J.; Song, H.; Dicks, E.; Tyrer, J.P.; Rosenthal, A.N.; Intermaggio, M.P.; Fraser, L.; Gentry-Maharaj, A.; Hayward, J.; Philpott, S.; et al. Germline Mutations in the BRIP1, BARD1, PALB2, and NBN Genes in Women With Ovarian Cancer. J. Natl. Cancer Inst. 2015, 107, djv214.
- Lu, H.M.; Li, S.; Black, M.H.; Lee, S.; Hoiness, R.; Wu, S.; Mu, W.; Huether, R.; Chen, J.; Sridhar, S.; et al. Association of Breast and Ovarian Cancers With Predisposition Genes Identified by Large-Scale Sequencing. JAMA Oncol 2019, 5, 51–57.
- Lilyquist, J.; LaDuca, H.; Polley, E.; Davis, B.T.; Shimelis, H.; Hu, C.; Hart, S.N.; Dolinsky, J.S.; Couch, F.J.; Goldgar, D.E. Frequency of mutations in a large series of clinically ascertained ovarian cancer cases tested on multi-gene panels compared to reference controls. Gynecol. Oncol. 2017, 147, 375–380.
- Norquist, B.M.; Harrell, M.I.; Brady, M.F.; Walsh, T.; Lee, M.K.; Gulsuner, S.; Bernards, S.S.; Casadei, S.; Yi, Q.; Burger, R.A.; et al. Inherited Mutations in Women With Ovarian Carcinoma. JAMA Oncol. 2016, 2, 482–490.
- Slavin, T.P.; Maxwell, K.N.; Lilyquist, J.; Vijai, J.; Neuhausen, S.L.; Hart, S.N.; Ravichandran, V.; Thomas, T.; Maria, A.; Villano, D.; et al. The contribution of pathogenic variants in breast cancer susceptibility genes to familial breast cancer risk. NPJ Breast Cancer 2017, 3, 22.
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1067.
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O'Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291.
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443.
- de Souza Timoteo, A.R.; Goncalves, A.; Sales, L.A.P.; Albuquerque, B.M.; de Souza, J.E.S.; de Moura, P.C.P.; de Aquino, M.A.A.; Agnez-Lima, L.F.; Lajus, T.B.P. A portrait of germline mutation in Brazilian at-risk for hereditary breast cancer. Breast Cancer Res. Treat. 2018, 172, 637–646.




