Authors: Piotr Kozlowski & Malwina Suszynska
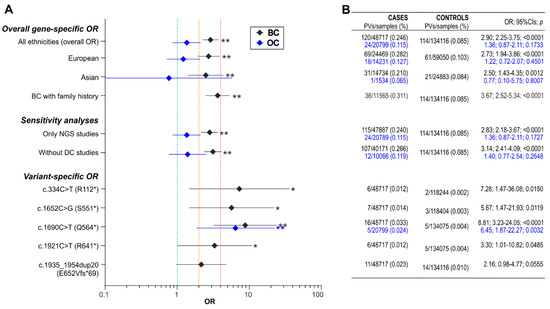
A catalog of BARD1 germline mutations/pathogenic variants (PVs) identified in large cumulative cohorts of ~48,700 breast cancer (BC) and ~20,800 ovarian cancer (OC) cases prepared based on 123 studies published over 20 years of BARD1 gene screening. By comparing the frequency of BARD1 PVs in the cases and ~134,100 noncancer controls from the gnomAD database, the effect of the BARD1 PVs on BC and OC risks is estimated.
- BARD1,
- mutation
- breast cancer
- ovarian cancer
- cancer risk
1. Definition
Over the last two decades, numerous BARD1 mutations/pathogenic variants (PVs) have been found in patients with breast cancer (BC) and ovarian cancer (OC). However, their role in BC and OC susceptibility remains controversial, and strong evidence-based guidelines for carriers are not yet available.
2. Introduction
3. BARD1 Mutational Spectrum
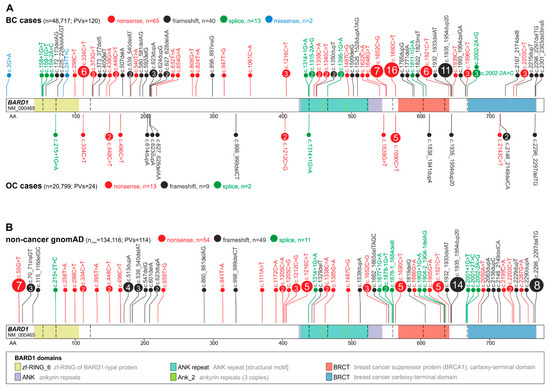
4. Association of BARD1 Pathogenic Variants with Breast Cancer
Funding
This work was supported by grants from the Polish National Science Centre (NCN 2015/17/B/NZ2/01182).Acknowledgments
The authors would like to thank the Genome Aggregation Database (gnomAD) and the groups that provided exome and genome variant data to this resource. A full list of contributing groups can be found at http://gnomad.broadinstitute.org/about.Author affiliations
Piotr Kozlowski & Malwina Suszynska, Department of Molecular Genetics, Institute of Bioorganic Chemistry, Polish Academy of Sciences, Noskowskiego 12/14 Street, 61-704, Poznan, PolandReferences
1.Suszynska, M.; Klonowska, K.; Jasinska, A.J.; Kozlowski, P. Large-scale meta-analysis of mutations identifiedin panels of breast/ovarian cancer-related genes—Providing evidence of cancer predisposition genes.Gynecol. Oncol.2019,153, 452–462.
2.Walsh, T.; Casadei, S.; Lee, M.K.; Pennil, C.C.; Nord, A.S.; Thornton, A.M.; Roeb, W.; Agnew, K.J.; Stray, S.M.;Wickramanayake, A.; et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritonealcarcinoma identified by massively parallel sequencing.Proc. Natl. Acad. Sci. USA2011,108, 18032–18037.
3.Couch, F.J.; Shimelis, H.; Hu, C.; Hart, S.N.; Polley, E.C.; Na, J.; Hallberg, E.; Moore, R.; Thomas, A.;Lilyquist, J.; et al. Associations Between Cancer Predisposition Testing Panel Genes and Breast Cancer.JAMA Oncol.2017,3, 1190–1196.
4.Daly, M.B.; Pilarski, R.; Yurgelun, M.B.; Berry, M.P.; Buys, S.S.; Dickson, P.; Domchek, S.M.; Elkhanany, A.;Friedman, S.; Garber, J.E.; et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast,Ovarian, and Pancreatic, Version 1.2020.J. Natl. Compr. Canc. Netw.2020,18, 380–391.
5.Daly, M.B.; Pilarski, R.; Berry, M.; Buys, S.S.; Farmer, M.; Friedman, S.; Garber, J.E.; Kauff, N.D.; Khan, S.;Klein, C.; et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast and Ovarian,Version 2.2017.J. Natl. Compr. Canc. Netw.2017,15, 9–20.
6.Hashizume, R.; Fukuda, M.; Maeda, I.; Nishikawa, H.; Oyake, D.; Yabuki, Y.; Ogata, H.; Ohta, T. The RINGheterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation.J. Biol. Chem.2001,276, 14537–14540.
7.Irminger-Finger, I.; Ratajska, M.; Pilyugin, M. New concepts on BARD1: Regulator of BRCA pathways andbeyond.Int. J. Biochem. Cell Biol.2016,72, 1–17.
8.Tarsounas, M.; Sung, P. The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication.Nat. Rev.Mol. Cell Biol.2020,21, 284–299.
9.Irminger-Finger, I.; Leung, W.C.; Li, J.; Dubois-Dauphin, M.; Harb, J.; Feki, A.; Jefford, C.E.; Soriano, J.V.;Jaconi, M.; Montesano, R.; et al. Identification of BARD1 as mediator between proapoptotic stress andp53-dependent apoptosis.Mol. Cell2001,8, 1255–1266.
10.Cimmino, F.; Formicola, D.; Capasso, M. Dualistic Role of BARD1 in Cancer.Genes2017,8, 375.
11.Li, L.; Ryser, S.; Dizin, E.; Pils, D.; Krainer, M.; Jefford, C.E.; Bertoni, F.; Zeillinger, R.; Irminger-Finger, I.Oncogenic BARD1 isoforms expressed in gynecological cancers.Cancer Res.2007,67, 11876–11885.
12.Zhang, Y.Q.; Bianco, A.; Malkinson, A.M.; Leoni, V.P.; Frau, G.; De Rosa, N.; Andre, P.A.; Versace, R.;Boulvain, M.; Laurent, G.J.; et al. BARD1: An independent predictor of survival in non-small cell lung cancer.Int. J. Cancer2012,131, 83–94.
13.Bosse, K.R.; Diskin, S.J.; Cole, K.A.; Wood, A.C.; Schnepp, R.W.; Norris, G.; Nguyen le, B.; Jagannathan, J.;Laquaglia, M.; Winter, C.; et al. Common variation at BARD1 results in the expression of an oncogenicisoform that influences neuroblastoma susceptibility and oncogenicity.Cancer Res.2012,72, 2068–2078.
14.Suszynska, M.; Kluzniak, W.; Wokolorczyk, D.; Jakubowska, A.; Huzarski, T.; Gronwald, J.; Debniak, T.;Szwiec, M.; Ratajska, M.; Klonowska, K.; et al. BARD1 is A Low/Moderate Breast Cancer Risk Gene: EvidenceBased on An Association Study of the Central European p.Q564X Recurrent Mutation.Cancers2019,11, 740.
15.Weber-Lassalle, N.; Borde, J.; Weber-Lassalle, K.; Horvath, J.; Niederacher, D.; Arnold, N.; Kaulfuss, S.;Ernst, C.; Paul, V.G.; Honisch, E.; et al. Germline loss-of-function variants in the BARD1 gene are associatedwith early-onset familial breast cancer but not ovarian cancer.Breast Cancer Res.2019,21, 55.
16.Ramus, S.J.; Song, H.; Dicks, E.; Tyrer, J.P.; Rosenthal, A.N.; Intermaggio, M.P.; Fraser, L.; Gentry-Maharaj, A.;Hayward, J.; Philpott, S.; et al. Germline Mutations in the BRIP1, BARD1, PALB2, and NBN Genes in WomenWith Ovarian Cancer.J. Natl. Cancer Inst.2015,107, djv214.
17.Lu, H.M.; Li, S.; Black, M.H.; Lee, S.; Hoiness, R.; Wu, S.; Mu, W.; Huether, R.; Chen, J.; Sridhar, S.; et al.Association of Breast and Ovarian Cancers With Predisposition Genes Identified by Large-Scale Sequencing.JAMA Oncol2019,5, 51–57. [CrossRef]
18.Lilyquist, J.; LaDuca, H.; Polley, E.; Davis, B.T.; Shimelis, H.; Hu, C.; Hart, S.N.; Dolinsky, J.S.; Couch, F.J.;Goldgar, D.E. Frequency of mutations in a large series of clinically ascertained ovarian cancer cases tested onmulti-gene panels compared to reference controls.Gynecol. Oncol.2017,147, 375–380.
19.Norquist, B.M.; Harrell, M.I.; Brady, M.F.; Walsh, T.; Lee, M.K.; Gulsuner, S.; Bernards, S.S.; Casadei, S.; Yi, Q.;Burger, R.A.; et al. Inherited Mutations in Women With Ovarian Carcinoma.JAMA Oncol.2016,2, 482–490.
20.Slavin, T.P.; Maxwell, K.N.; Lilyquist, J.; Vijai, J.; Neuhausen, S.L.; Hart, S.N.; Ravichandran, V.; Thomas, T.;Maria, A.; Villano, D.; et al. The contribution of pathogenic variants in breast cancer susceptibility genes tofamilial breast cancer risk.NPJ Breast Cancer2017,3, 22.
21.Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.;Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456humans.Nature2020,581, 434–443.
22.Tung, N.; Battelli, C.; Allen, B.; Kaldate, R.; Bhatnagar, S.; Bowles, K.; Timms, K.; Garber, J.E.; Herold, C.;Ellisen, L.; et al. Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2testing using next-generation sequencing with a 25-gene panel.Cancer2015,121, 25–33.
23.Susswein, L.R.; Marshall, M.L.; Nusbaum, R.; Vogel Postula, K.J.; Weissman, S.M.; Yackowski, L.; Vaccari, E.M.;Bissonnette, J.; Booker, J.K.; Cremona, M.L.; et al. Pathogenic and likely pathogenic variant prevalenceamong the first 10,000 patients referred for next-generation cancer panel testing.Genet. Med.2016,18,823–832.
24.LaDuca, H.; Stuenkel, A.J.; Dolinsky, J.S.; Keiles, S.; Tandy, S.; Pesaran, T.; Chen, E.; Gau, C.L.; Palmaer, E.;Shoaepour, K.; et al. Utilization of multigene panels in hereditary cancer predisposition testing: Analysis ofmore than 2,000 patients.Genet. Med.2014,16, 830–837.
