
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | M. Catarina M.D. De Almeida | + 2282 word(s) | 2282 | 2021-09-15 06:34:12 | | | |
| 2 | Lindsay Dong | Meta information modification | 2282 | 2021-09-22 04:05:35 | | |
Video Upload Options
Some wild-type and genetically modified bacteria can metabolize xylose through three different main pathways of metabolism: xylose isomerase pathway, oxidoreductase pathway, and non-phosphorylative pathway (including Weimberg and Dahms pathways). Two of the commercially interesting intermediates of these pathways are xylitol and xylonic acid, which can accumulate in the medium either through manipulation of the culture conditions or through genetic modification of the bacteria.
1. Introduction
2. Major Routes of Xylose Transport and Metabolism in Bacteria
Xylose is a very common sugar in residual lignocellulosic biomass, being the second major sugar found in most lignocellulosic hydrolysates and the major sugar in hemicellulosic hydrolysates. For that reason, xylose is a very promising carbon source, and it makes sense to understand the fundamentals of the mechanism used by bacteria to metabolize xylose into high-value by-products.
2.1. Mechanisms of Xylose Transport
Regarding D-xylose, yeast and fungi can use facilitated diffusion or active transport, while bacteria tend to use active transport mechanisms [6]. These types of mechanisms are mediated by carrier proteins and, hence, exhibit the properties of specific inhibition, substrate specificity, and saturability. These processes enable sugar transportation against a concentration gradient at the expense of metabolic energy.
Bacteria species such as Bacillus, Clostridium, Escherichia coli, and Lactococcus use active transport for the uptake of xylose into the cell [6][7]. Usually, there are high- and low-affinity transporter routes. In E. coli, the most studied species, the high-affinity transporter (XylFGH) belongs to the ATP-binding cassette (ABC) family, while the low-affinity transporter (XlyE), a proton-coupled symporter, belongs to the major facilitator superfamily (MFS), with xylose transport being driven by a proton motive force [8][9][10][11]. The low-affinity transport mechanism is also present in Bacillus megaterium, Bacillus subtilis, Lactobacillus brevis, Salmonella typhimurium, Tetragenococcus halophila, and some ruminal bacteria [10][12][13][14][15][16][17][18]. The ABC transporter is more efficient concerning xylose uptake and comprises a D-xylose-binding protein XylF, the membrane permease XylH, and the ATP-binding protein XylG [11][19]. This system is also present in bacteria such as Clostridium, Geobacillus, or Thermoanaerobacter species [6][20][21][22]. However, this transport can be inhibited when other readily fermentable substrates, such as glucose, are present. Many microbial strains have a regulatory mechanism, carbon catabolite repression (CCR), mainly mediated by components of the phosphoenolpyruvate (PEP): carbohydrate phosphotransferase system (PTS), which prevents the expression of genes needed for catabolism of other carbon sources, namely pentoses, while the substrate that enables the fastest growth (normally glucose) is present [23][24]. Concerning E. coli, at least two mechanisms of xylose transport and metabolism repression were reported, including the XylR regulator and cyclic AMP (cAMP) receptor protein (CRP)-dependent control of Xyl genes and the presence of arabinose, since the transporters allow for the transportation of this sugar at lower efficiencies [25]. The presence of high levels of glucose leads to the dephosphorylation of the component EIIA of PTS, which becomes unable to activate the enzyme adenylate cyclase, originating low levels of cAMP. In contrast, when glucose levels drop, cAMP increases, activating CRP that, together with XylR (activated when bound by xylose), stimulate the operons xylFGH and xylAB, involved in xylose transport and metabolism [16]. These repressive mechanisms could bring difficulties in the utilization of lignocellulosic hydrolysates, where both sugars are present. In this kind of media, a diauxic growth is observed and the preferential substrate, usually glucose, is consumed first. When glucose is depleted in the culture medium and another non-repressive substrate such as xylose is present, there is a temporary cessation of growth and catabolic repression is then relieved.
2.2. Xylose Metabolic Network in Bacteria
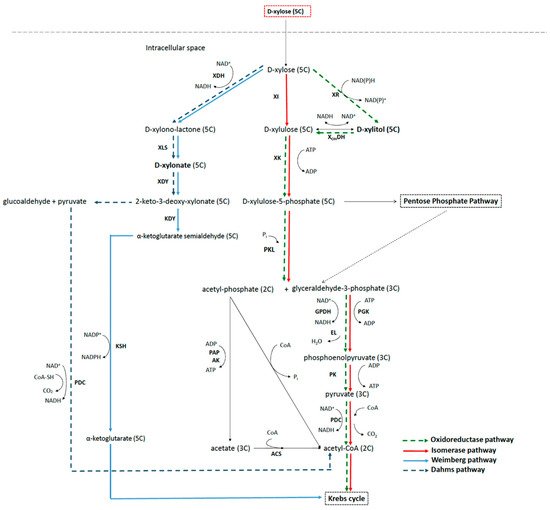
2.3. Metabolic Pathways to Xylitol and Xylonic Acid
2.3.1. Xylitol
Table 1 summarizes the contents of this section regarding a selection of bacterial strains able to convert xylose into xylitol.
| Strains | C-Source | Genetic Modification | Growth Conditions | Xylitol (g L−1) * |
Yxylitol/xylose (g g−1) |
Productivity (g L−1 h−1) * |
Ref. |
|---|---|---|---|---|---|---|---|
| Corynebacterium glutamicum Cg-ax3 |
arabinose glucose xylose |
Yes | Batch shake flask | 6.7 | n.a. | n.a. | [47] |
| Fed-batch shake flask | 31 | n.a. | 0.28 gg−1cdw h−1 | ||||
| acid pre-treated liquor of sorghum stover | Fed-batch shake flask | 27 | n.a. | 0.22 gg−1cdw h−1 | |||
| Corynebacterium sp. NRRL B 4247 | xylose | No | Shake flask | 1.7 | 0.57 | 0.071 | [48] |
| 6-phosphogluconate (source of NADPH) added to the medium Shake flask |
10 | n.a. | 0.067 | ||||
| Corynebacterium sp. no. 208 | xylose | No | 6-phosphogluconate (source of NADPH) was added to the medium Shake flask |
69 | n.a. | 0.21 | [49] |
| Enterobacter liquefaciens 553 |
xylose | No | Shake flask | 33 | n.a. | 0.35 | [50] |
| E. coli BL21(DE3) | xylose | Yes | Shake flask | 202 | 1.0 | 6.37 | [51] |
| Escherichia coli IS5-d | xylose and glucose | Yes | 5 L Batch STR | 110 | n.a. | 3.06 | [52] |
| Escherichia coli IS5-M | corncob hemicellulosic hydrolysate and 24 g L−1 corn steep liquor | Yes | 15 L Fed-batch STR | 144 | n.a. | 1.84 | [52] |
| Escherichia coli HK402 | xylose and glucose | Yes | 15 L Fed-batch STR | 172 | >0.95 | 1.57 | [52] |
| detoxified hemicellulosic hydrolysate and glucose | 150 | >0.95 | 1.40 | ||||
| Escherichia coli WZ51 | detoxified hemicellulosic hydrolysate | Yes | 15 L Fed-batch STR | 132 | 0.95 | 2.09 | [53] |
| Mycobacterium smegmatis | xylose | No | immobilized D-xylose isomerase from Bacillus coagulans and immobilized M. smegmatis Shake flask |
5 g | 0.80 | n.a. | [54] |
| Paraburkholderia sacchari DSM 17165 |
xylose | No | 2 L Fed-batch STR | 17 | n.a. | 0.39 | [23] |
| Paraburkholderia sacchari DSM 17165 |
xylose | No | 2 L Fed-batch STR | 70 | 0.39 | 0.50 | [31] |
2.3.2. Xylonic Acid
Table 2 gathers information on various examples of bacterial strains able to convert xylose into xylonic acid.
| Strains | C-Source | Genetic Modification | Growth Conditions | Xylonic Acid (g L−1) * |
Yxylonic acid/xylose (g g−1) |
Productivity (g L−1 h−1) * |
Ref. |
|---|---|---|---|---|---|---|---|
| Corynebacterium glutamicum ATCC13032 | xylose | Yes | Shake flask | 50.7 | 0.76 | 0.42 | [55] |
| Corynebacterium glutamicum ATCC31831 | rice straw hydrolysate after dilute sulfuric acid pretreatment | Yes | Shake flask | 42.9 | 1.1 | 0.37 | [55] |
| xylose | 56.3 | 0.84 | 0.47 | ||||
| Escherichia coli BL21 |
xylose | Yes | Shake flask | 9.1 | 1.10 | 0.45 | [56] |
| 2 L Batch STR | 6.9 | 0.89 | 0.11 | ||||
| Escherichia coli W3110 |
xylose and glucose | Yes | Shake flask | 5.1 | 0.51 | 0.084 | [57] |
| 5 L Fed-batch STR | 39.2 | 0.98 | 1.09 | ||||
| Escherichia coli BL21 |
xylose and glycerol | Yes | 5 L Fed-batch STR | 27.3 | n.a. | 1.8 | [58] |
| Gluconobacter oxydans ATCC 621 |
xylose | No | 3 L Batch STR | 109 | 0.95 | 2.5 | [59] |
| steamed and enzymatically hydrolyzed birchwood | 12.4 | 0.50 | n.a. | ||||
| Gluconobacter oxydans DSM 2003 |
corn stover hydrolysate after dry dilute acid pretreatment | No | 3 L Batch STR | 38.9 | 0.9 | n.a. | [60] |
| Gluconobacter oxydans DSM 2003 |
xylose | No | 3 L Batch STR | 66.4 | n.a. | 5.5 | [61] |
| Gluconobacter oxydans NL71 |
xylose | No | Compressed oxygen-supplied sealed stirred tank reactor (COS-SSTR); pure oxygen supply | 586.3 | 0.95 | 4.7 | [62] |
| corn stover diluted sulfuric acid hydrolysates without detoxification | 143.9 | 0.97 | 1.0 | ||||
| Gluconobacter oxydans NL71 |
xylose in distillation stillage of cellulosic ethanol fermentation broth |
No | COS-SSTR; fed-batch addition of xylose with cell-recycling | 1813 g in 6-fold cell recycling; 1 L culture medium | n.a. | 16.8 g h−1 in 108 h | [63] |
| Gluconobacter oxydans NL71 |
corn stover hydrolysate after dry diluted acid pretreatment | No | Two-stage fermentation in a 3 L COS-SSTR bioreactor with cell recycling | 167.4 g from 1 kg corn stover | 0.97 | 3.7 | [64] |
| Gluconobacter oxydans ATCC 621 |
xylose | No | Fed-batch bioreactor; Immobilized whole-cells; pressurized pure oxygen supply followed by electrodialysis acid chamber (POA-SSB-OE) | 329.2 g xylonic acid | n.a. | 7.1 g h−1 in 48 h | [65] |
| Klebsiella pneumoniae (modified) |
bamboo hydrolysate | Yes | Fed-batch cultivations | 103 | 0.98 | n.a. | [66] |
| Paraburkholderia sacchari DSM 17165 |
xylose | No | 2 L Fed-batch STR xylose as carbon source; high dissolved oxygen concentration |
150 | 0.85 | 1.5 | [31] |
| xylose and glucose | 2 L Fed-batch STR high dissolved oxygen concentration |
390 | 1.1 | 6.0 |
3. Conclusions
References
- Kamm, B.; Kamm, M. Principles of biorefineries. Appl. Microbiol. Biotechnol. 2004, 64, 137–145.
- Ferreira, A.F. Biorefinery concept. In Lecture Notes in Energy—Biorefineries: Targeting Energy, High Value Products and Waste Valorisation; Rabaçal, M., Ferreira, A.F., Silva, C.A.M., Costa, M., Eds.; Springer International Publishing AG: Cham, Switzerland, 2017; pp. 1–20. ISBN 978-3-319-48286-6.
- Cherubini, F. The biorefinery concept: Using biomass instead of oil for producing energy and chemicals. Energy Convers. Manag. 2010, 51, 1412–1421.
- Moncada, J.; Aristizábal, V. Design strategies for sustainable biorefineries. Biochem. Eng. J. 2016, 116, 122–134.
- Irmak, S. Biomass as Raw Material for Production of High-Value Products. In Biomass Volume Estimation and Valorization for Energy; Tumuluru, J.S., Ed.; IntechOpen: Rijeka, Croatia, 2017; pp. 201–225. ISBN 978-953-51-2938-7.
- McClintock, M.K.; Zhang, K. Xylose Metabolism and Its Metabolic Engineering Applications. In Engineering Microbial Metabolism for Chemical Synthesis; World Scientific (Europe): Hackensack, NJ, USA, 2017; pp. 209–235. ISBN 978-1-78634-429-8.
- Gu, Y.; Ding, Y.; Ren, C.; Sun, Z.; Rodionov, D.A.; Zhang, W.; Yang, S.; Yang, C.; Jiang, W. Reconstruction of xylose utilization pathway and regulons in Firmicutes. BMC Genom. 2010, 11, 255.
- Davis, E.O.; Henderson, P.J. The cloning and DNA sequence of the gene xylE for xylose-proton symport in Escherichia coli K12. J. Biol. Chem. 1987, 262, 13928–13932.
- Khankal, R.; Chin, J.W.; Cirino, P.C. Role of xylose transporters in xylitol production from engineered Escherichia coli. J. Biotechnol. 2008, 134, 246–252.
- Shamanna, D.K.; Sanderson, K.E. Uptake and catabolism of D-xylose in Salmonella typhimurium LT2. J. Bacteriol. 1979, 139, 64–70.
- Song, S.; Park, C. Organization and regulation of the D-xylose operons in Escherichia coli K-12: XylR acts as a transcriptional activator. J. Bacteriol. 1997, 179, 7025–7032.
- Chaillou, S.; Bor, Y.C.; Batt, C.A.; Postma, P.W.; Pouwels, P.H. Molecular cloning and functional expression in lactobacillus plantarum 80 of xylT, encoding the D-xylose-H+ symporter of Lactobacillus brevis. Appl. Environ. Microbiol. 1998, 64, 4720–4728.
- Chaillou, S.; Pouwels, P.H.; Postma, P.W. Transport of D-Xylose in Lactobacillus pentosus, Lactobacillus casei, and Lactobacillus plantarum: Evidence for a Mechanism of Facilitated Diffusion via the Phosphoenolpyruvate:Mannose Phos. J. Bacteriol. 1999, 181, 4768–4773.
- Schmiedel, D.; Hillen, W. A Bacillus subtilis 168 mutant with increased xylose uptake can utilize xylose as sole carbon source. FEMS Microbiol. Lett. 1996, 135, 175–178.
- Schmiedel, D.; Kintrup, M.; Küster, E.; Hillen, W. Regulation of expression, genetic organization and substrate specificity of xylose uptake in Bacillus megaterium. Mol. Microbiol. 1997, 23, 1053–1062.
- Sievert, C.; Nieves, L.M.; Panyon, L.A.; Loeffler, T.; Morris, C.; Cartwright, R.A.; Wang, X. Experimental evolution reveals an effective avenue to release catabolite repression via mutations in XylR. Proc. Natl. Acad. Sci. USA 2017, 114, 7349–7354.
- Strobel, H.J. Evidence for catabolite inhibition in regulation of pentose utilization and transport in the ruminal bacterium Selenomonas ruminantium. Appl. Environ. Microbiol. 1993, 59, 40–46.
- Takeda, Y.; Takase, K.; Yamato, I.; Abe, K. Sequencing and characterization of the xyl operon of a gram-positive bacterium, Tetragenococcus halophila. Appl. Environ. Microbiol. 1998, 64, 2513–2519.
- Li, J.; Wang, C.; Yang, G.; Sun, Z.; Guo, H.; Shao, K.; Gu, Y.; Jiang, W.; Zhang, P. Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria. Proc. Natl. Acad. Sci. USA 2017, 114, 8235–8240.
- Erbeznik, M.; Hudson, S.E.; Herrman, A.B.; Strobel, H.J. Molecular analysis of the xylFGH operon, coding for xylose ABC transport, in Thermoanaerobacter ethanolicus. Curr. Microbiol. 2004, 48, 295–299.
- Shulami, S.; Zaide, G.; Zolotnitsky, G.; Langut, Y.; Feld, G.; Sonenshein, A.L.; Shoham, Y. A two-component system regulates the expression of an ABC transporter for xylo-oligosaccharides in Geobacillus stearothermophilus. Appl. Environ. Microbiol. 2007, 73, 874–884.
- Sun, Z.; Chen, Y.; Yang, C.; Yang, S.; Gu, Y.; Jiang, W. A novel three-component system-based regulatory model for D-xylose sensing and transport in Clostridium beijerinckii. Mol. Microbiol. 2015, 95, 576–589.
- Raposo, R.S.; de Almeida, M.C.; de Oliveira, M.D.C.; da Fonseca, M.M.; Cesário, M.T. A Burkholderia sacchari cell factory: Production of poly-3-hydroxybutyrate, xylitol and xylonic acid from xylose-rich sugar mixtures. N. Biotechnol. 2017.
- Wang, X.; Goh, E.-B.; Beller, H.R. Engineering, E. coli for simultaneous glucose–xylose utilization during methyl ketone production. Microb. Cell Fact. 2018, 17, 12.
- Desai, T.A.; Rao, C. V Regulation of arabinose and xylose metabolism in Escherichia coli. Appl. Environ. Microbiol. 2010, 76, 1524–1532.
- Lawlis, V.B.; Dennis, M.S.; Chen, E.Y.; Smith, D.H.; Henner, D.J. Cloning and sequencing of the xylose isomerase and xylulose kinase genes of Escherichia coli. Appl. Environ. Microbiol. 1984, 47, 15–21.
- Lokman, B.C.; van Santen, P.; Verdoes, J.C.; Krüse, J.; Leer, R.J.; Posno, M.; Pouwels, P.H. Organization and characterization of three genes involved in D-xylose catabolism in Lactobacillus pentosus. Mol. Gen. Genet. 1991, 230, 161–169.
- Rygus, T.; Scheler, A.; Allmansberger, R.; Hillen, W. Molecular cloning, structure, promoters and regulatory elements for transcription of the Bacillus megaterium encoded regulon for xylose utilization. Arch. Microbiol. 1991, 155, 535–542.
- Stephens, C.; Christen, B.; Fuchs, T.; Sundaram, V.; Watanabe, K.; Jenal, U. Genetic analysis of a novel pathway for D-xylose metabolism in Caulobacter crescentus. J. Bacteriol. 2007, 189, 2181–2185.
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999, 27, 29–34.
- Bondar, M.; da Fonseca, M.M.R.; Cesário, M.T. Xylonic acid production from xylose by Paraburkholderia sacchari. Biochem. Eng. J. 2021, 170, 107982.
- Wong, H.C.; Ting, Y.; Lin, H.C.; Reichert, F.; Myambo, K.; Watt, K.W.; Toy, P.L.; Drummond, R.J. Genetic organization and regulation of the xylose degradation genes in Streptomyces rubiginosus. J. Bacteriol. 1991, 173, 6849–6858.
- Yamanaka, K. Inhibition of d-xylose isomerase by pentitols and d-lyxose. Arch. Biochem. Biophys. 1969, 131, 502–506.
- Kovalevsky, A.; Hanson, B.L.; Mason, S.A.; Forsyth, V.T.; Fisher, Z.; Mustyakimov, M.; Blakeley, M.P.; Keen, D.A.; Langan, P. Inhibition of D-xylose isomerase by polyols: Atomic details by joint X-ray/neutron crystallography. Acta Crystallogr. Sect. D 2012, 68, 1201–1206.
- Moat, A.G.; Foster, J.W.; Spector, M.P. Microbial Physiology, 4th ed.; Wiley-Liss, Inc.: New York, NY, USA, 2002; ISBN 0-471-39483-1.
- Gu, Y.; Jiang, Y.; Yang, S.; Jiang, W. Utilization of economical substrate-derived carbohydrates by solventogenic clostridia: Pathway dissection, regulation and engineering. Curr. Opin. Biotechnol. 2014, 29, 124–131.
- Liu, L.; Zhang, L.; Tang, W.; Gu, Y.; Hua, Q.; Yang, S.; Jiang, W.; Yang, C. Phosphoketolase pathway for Xylose Catabolism in Clostridium acetobutylicum revealed by 13C metabolic flux analysis. J. Bacteriol. 2012, 194, 5413–5422.
- Tanaka, K.; Komiyama, A.; Sonomoto, K.; Ishizaki, A.; Hall, S.; Stanbury, P. Two different pathways for D-xylose metabolism and the effect of xylose concentration on the yield coefficient of L-lactate in mixed-acid fermentation by the lactic acid bacterium Lactococcus lactis IO-1. Appl. Microbiol. Biotechnol. 2002, 60, 160–167.
- Kwak, S.; Jin, Y.-S. Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: A review and perspective. Microb. Cell Fact. 2017, 16, 82.
- McClintock, M.K.; Wang, J.; Zhang, K. Application of Nonphosphorylative Metabolism as an Alternative for Utilization of Lignocellulosic Biomass. Front. Microbiol. 2017, 8, 2310.
- Weimberg, R. Pentose oxidation by Pseudomonas fragi. J. Biol. Chem. 1961, 236, 629–635.
- Köhler, K.A.K.; Blank, L.M.; Frick, O.; Schmid, A. D-Xylose assimilation via the Weimberg pathway by solvent-tolerant Pseudomonas taiwanensis VLB120. Environ. Microbiol. 2015, 17, 156–170.
- Franden, M.A.; Jayakody, L.N.; Li, W.-J.; Wagner, N.J.; Cleveland, N.S.; Michener, W.E.; Hauer, B.; Blank, L.M.; Wierckx, N.; Klebensberger, J.; et al. Engineering Pseudomonas putida KT2440 for efficient ethylene glycol utilization. Metab. Eng. 2018, 48, 197–207.
- Nunn, C.E.M.; Johnsen, U.; Schönheit, P.; Fuhrer, T.; Sauer, U.; Hough, D.W.; Danson, M.J. Metabolism of pentose sugars in the hyperthermophilic archaea Sulfolobus solfataricus and Sulfolobus acidocaldarius. J. Biol. Chem. 2010, 285, 33701–33709.
- Bañares, A.B.; Nisola, G.M.; Valdehuesa, K.N.G.; Lee, W.K.; Chung, W.J. Understanding D-xylonic acid accumulation: A cornerstone for better metabolic engineering approaches. Appl. Microbiol. Biotechnol. 2021, 105, 5309–5324.
- Kim, D.; Woo, H.M. Deciphering bacterial xylose metabolism and metabolic engineering of industrial microorganisms for use as efficient microbial cell factories. Appl. Microbiol. Biotechnol. 2018, 102, 9471–9480.
- Dhar, K.S.; Wendisch, V.F.; Nampoothiri, K.M. Engineering of Corynebacterium glutamicum for xylitol production from lignocellulosic pentose sugars. J. Biotechnol. 2016, 230, 63–71.
- Rangaswamy, S.; Agblevor, F.A. Screening of facultative anaerobic bacteria utilizing D-xylose for xylitol production. Appl. Microbiol. Biotechnol. 2002, 60, 88–93.
- Yoshitake, J.; Ohiwa, H.; Shimamura, M.; Imai, T. Production of Polyalcohol by a Corynebacterium sp. Agric. Biol. Chem. 1971, 35, 905–911.
- Yoshitake, J.; Ishizaki, H.; Shimamura, M.; Imai, T. Xylitol Production by an Enterobacter Species. Agric. Biol. Chem. 1973, 37, 2261–2267.
- Jin, L.-Q.; Xu, W.; Yang, B.; Liu, Z.-Q.; Zheng, Y.-G. Efficient Biosynthesis of Xylitol from Xylose by Coexpression of Xylose Reductase and Glucose Dehydrogenase in Escherichia coli. Appl. Biochem. Biotechnol. 2019, 187, 1143–1157.
- Su, B.; Zhang, Z.; Wu, M.; Lin, J.; Yang, L. Construction of plasmid-free Escherichia coli for the production of arabitol-free xylitol from corncob hemicellulosic hydrolysate. Sci. Rep. 2016, 6, 26567.
- Yuan, X.; Wang, J.; Lin, J.; Yang, L.; Wu, M. Efficient production of xylitol by the integration of multiple copies of xylose reductase gene and the deletion of Embden–Meyerhof–Parnas pathway-associated genes to enhance NADPH regeneration in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2019, 46, 1061–1069.
- Izumori, K.; Tuzaki, K. Production of xylitol from D-xylulose by Mycobacterium smegmatis. J. Ferment. Technol. 1988, 66, 33–36.
- Sundar, M.S.L.; Susmitha, A.; Rajan, D.; Hannibal, S.; Sasikumar, K.; Wendisch, V.F.; Nampoothiri, K.M. Heterologous expression of genes for bioconversion of xylose to xylonic acid in Corynebacterium glutamicum and optimization of the bioprocess. AMB Express 2020, 10, 68.
- Rodzri, N.A.M.; Zain, W.S.; Hanapiah, R.M.A.; Samah, R.A.; Illias, R. D-Xylonic Acid from Recombinant E. coli BL21 (DE3): Comparison between Shake Flask and Benchtop Bioreactor Fermentation. In Journal of Physics: Conference Series; IOP Publishing: Bristol, UK, 2020.
- Liu, H.; Valdehuesa, K.N.G.; Nisola, G.M.; Ramos, K.R.M.; Chung, W.-J. High yield production of d-xylonic acid from d-xylose using engineered Escherichia coli. Bioresour. Technol. 2012, 115, 244–248.
- Cao, Y.; Xian, M.; Zou, H.; Zhang, H. Metabolic engineering of Escherichia coli for the production of xylonate. PLoS ONE 2013, 8, e67305.
- Buchert, J.; Puls, J.; Poutanen, K. Comparison of Pseudomonas fragi and Gluconobacter oxydans for production of xylonic acid from hemicellulose hydrolyzates. Appl. Microbiol. Biotechnol. 1988, 28, 367–372.
- Zhang, H.; Liu, G.; Zhang, J.; Bao, J. Fermentative production of high titer gluconic and xylonic acids from corn stover feedstock by Gluconobacter oxydans and techno-economic analysis. Bioresour. Technol. 2016, 219, 123–131.
- Zhang, H.; Han, X.; Wei, C.; Bao, J. Oxidative production of xylonic acid using xylose in distillation stillage of cellulosic ethanol fermentation broth by Gluconobacter oxydans. Bioresour. Technol. 2017, 224, 573–580.
- Zhou, X.; Lü, S.; Xu, Y.; Mo, Y.; Yu, S. Improving the performance of cell biocatalysis and the productivity of xylonic acid using a compressed oxygen supply. Biochem. Eng. J. 2015, 93, 196–199.
- Zhou, X.; Zhou, X.; Xu, Y. Improvement of fermentation performance of Gluconobacter oxydans by combination of enhanced oxygen mass transfer in compressed-oxygen-supplied sealed system and cell-recycle technique. Bioresour. Technol. 2017, 244, 1137–1141.
- Zhou, X.; Zhou, X.; Liu, G.; Xu, Y.; Balan, V. Integrated production of gluconic acid and xylonic acid using dilute acid pretreated corn stover by two-stage fermentation. Biochem. Eng. J. 2018, 137, 18–22.
- Zhou, X.; Han, J.; Xu, Y. Electrodialytic bioproduction of xylonic acid in a bioreactor of supplied-oxygen intensification by using immobilized whole-cell Gluconobacter oxydans as biocatalyst. Bioresour. Technol. 2019, 282, 378–383.
- Wang, C.; Wei, D.; Zhang, Z.; Wang, D.; Shi, J.; Kim, C.H.; Jiang, B.; Han, Z.; Hao, J. Production of xylonic acid by Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 2016, 100, 10055–10063.




