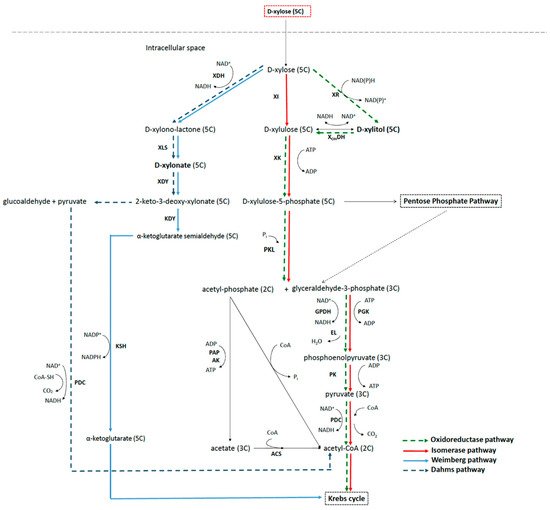
Some wild-type and genetically modified bacteria can metabolize xylose through three different main pathways of metabolism: xylose isomerase pathway, oxidoreductase pathway, and non-phosphorylative pathway (including Weimberg and Dahms pathways). Two of the commercially interesting intermediates of these pathways are xylitol and xylonic acid, which can accumulate in the medium either through manipulation of the culture conditions or through genetic modification of the bacteria.
1. Introduction
A great fraction of worldwide energy and material products come from fossil fuel refineries. The environmental concerns raised by the exploitation of fossil resources due to excessive pollution and consequent global warming, their ongoing price increase, uncertain availability, and non-sustainability are seen as reasons to invest in alternative solutions able to mitigate climate change and to reduce the consumption of fossil fuels.
The replacement of oil with biomass as raw material for fuel and chemical production is an interesting option and the driving force for the development of biorefinery plants, which operate in a similar way to oil refineries. In fact, the biorefinery concept can be explained as the processing of renewable biomass into a spectrum of fuels and valuable products [1][2][3][4]. This strategy needs a large investment to achieve the sustainability goals of our society. The replacement of oil with biomass as raw material for fuel and chemical production is an interesting option and the driving force for the development of biorefinery plants, which operate in a similar way to oil refineries. In fact, the biorefinery concept can be explained as the processing of renewable biomass into a spectrum of fuels and valuable products [6,8,11,12]. This strategy needs a large investment to achieve the sustainability goals of our society.
One challenge that is common to all industries and crucial to the success of the business is to ensure abundant and inexpensive raw materials in order to manufacture products with a competitive market price. In the case of bioprocesses, the raw material in question is carbon sources. Biomass and biomass-derived materials have been pointed out as one of the most promising alternatives. Biomass, in this context, refers to a rich carbon renewable raw material that can substitute fossil-based raw materials in the energy and chemical products industries. In biorefinery, almost all types of biomass that come from forestry residues, marine plants, waste food crops, food processing, animal farming, or human wastes can be converted to different classes of biofuels and biochemicals through jointly applied conversion technologies [1][5]. These products can be intermediates and/or final compounds in food, feed, materials, chemicals, and energy production (fuels, power, and heat) [4]. Therefore, biomass is considered the only sustainable source of organic carbon and the perfect equivalent to petroleum for the production of fuels and chemicals. One challenge that is common to all industries and crucial to the success of the business is to ensure abundant and inexpensive raw materials in order to manufacture products with a competitive market price. In the case of bioprocesses, the raw material in question is carbon sources. Biomass and biomass-derived materials have been pointed out as one of the most promising alternatives. Biomass, in this context, refers to a rich carbon renewable raw material that can substitute fossil-based raw materials in the energy and chemical products industries. In biorefinery, almost all types of biomass that come from forestry residues, marine plants, waste food crops, food processing, animal farming, or human wastes can be converted to different classes of biofuels and biochemicals through jointly applied conversion technologies [6,13]. These products can be intermediates and/or final compounds in food, feed, materials, chemicals, and energy production (fuels, power, and heat) [12]. Therefore, biomass is considered the only sustainable source of organic carbon and the perfect equivalent to petroleum for the production of fuels and chemicals.
2. Major Routes of Xylose Transport and Metabolism in Bacteria
Xylose is a very common sugar in residual lignocellulosic biomass, being the second major sugar found in most lignocellulosic hydrolysates and the major sugar in hemicellulosic hydrolysates. For that reason, xylose is a very promising carbon source, and it makes sense to understand the fundamentals of the mechanism used by bacteria to metabolize xylose into high-value by-products.
2.1. Mechanisms of Xylose Transport
Regarding D-xylose, yeast and fungi can use facilitated diffusion or active transport, while bacteria tend to use active transport mechanisms [6][41]. These types of mechanisms are mediated by carrier proteins and, hence, exhibit the properties of specific inhibition, substrate specificity, and saturability. These processes enable sugar transportation against a concentration gradient at the expense of metabolic energy.
Bacteria species such as BacillusBacillus, Clostridium, Escherichia coli, Clostridium, Escherichia coli, and Lactococcus use active transport for the uptake of xylose into the cell [6][7][41,44]. Usually, there are high- and low-affinity transporter routes. In E. coli, the most studied species, the high-affinity transporter (XylFGH) belongs to the ATP-binding cassette (ABC) family, while the low-affinity transporter (XlyE), a proton-coupled symporter, belongs to the major facilitator superfamily (MFS), with xylose transport being driven by a proton motive force [8][9][10][11][45,46,47,48]. The low-affinity transport mechanism is also present in Bacillus megaterium, Bacillus subtilis, Lactobacillus brevis, Salmonella typhimurium, Tetragenococcus halophila, and some ruminal bacteria [10][12][13][14][15][16][17][18][47,49,50,51,52,53,54,55]. The ABC transporter is more efficient concerning xylose uptake and comprises a D-xylose-binding protein XylF, the membrane permease XylH, and the ATP-binding protein XylG [11][19][48,56]. This system is also present in bacteria such as Clostridium, Geobacillus, or ThermoanaerobacterThermoanaerobacter species species [6][20][21][22][41,57,58,59]. However, this transport can be inhibited when other readily fermentable substrates, such as glucose, are present. Many microbial strains have a regulatory mechanism, carbon catabolite repression (CCR), mainly mediated by components of the phosphoenolpyruvate (PEP): carbohydrate phosphotransferase system (PTS), which prevents the expression of genes needed for catabolism of other carbon sources, namely pentoses, while the substrate that enables the fastest growth (normally glucose) is present [23][24][60,61]. Concerning E. coli, at least two mechanisms of xylose transport and metabolism repression were reported, including the XylR regulator and cyclic AMP (cAMP) receptor protein (CRP)-dependent control of Xyl genes and the presence of arabinose, since the transporters allow for the transportation of this sugar at lower efficiencies [25][62]. The presence of high levels of glucose leads to the dephosphorylation of the component EIIA of PTS, which becomes unable to activate the enzyme adenylate cyclase, originating low levels of cAMP. In contrast, when glucose levels drop, cAMP increases, activating CRP that, together with XylR (activated when bound by xylose), stimulate the operons xylFGH and xylAB, involved in xylose transport and metabolism [16][53]. These repressive mechanisms could bring difficulties in the utilization of lignocellulosic hydrolysates, where both sugars are present. In this kind of media, a diauxic growth is observed and the preferential substrate, usually glucose, is consumed first. When glucose is depleted in the culture medium and another non-repressive substrate such as xylose is present, there is a temporary cessation of growth and catabolic repression is then relieved.
2.2. Xylose Metabolic Network in Bacteria
3.2. Xylose Metabolic Network in Bacteria
Xylose is consumed mainly through three different metabolic pathways: (i) the isomerase pathway; (ii) the oxidoreductase pathway; and (iii) the oxidative pathway, also recognized as the non-phosphorylative pathway. The xylose metabolic network scheme is represented in
Figure 1. The isomerase pathway converts xylose into xylulose, which is then phosphorylated to xylulose phosphate entering the pentose phosphate pathway (PPP). This is typically used by prokaryotes, namely by E. coli, Bacillus sp., and Lactobacillus sp. [26][27][28][29]; the oxidoredutase pathway is mostly present in eukaryotic microorganisms. Here, xylose is first converted to xylitol, which is then dehydrogenated and phosphorylated to xylulose phosphate, entering the PPP. The oxidative pathway has a common trunk that divides in two branches: the Weimberg and the Dahms pathways. This oxidative pathway is mainly carried out by bacteria. . The isomerase pathway converts xylose into xylulose, which is then phosphorylated to xylulose phosphate entering the pentose phosphate pathway (PPP). This is typically used by prokaryotes, namely by E. coli, Bacillus sp., and Lactobacillus sp. [75,76,77,78]; the oxidoredutase pathway is mostly present in eukaryotic microorganisms. Here, xylose is first converted to xylitol, which is then dehydrogenated and phosphorylated to xylulose phosphate, entering the PPP. The oxidative pathway has a common trunk that divides in two branches: the Weimberg and the Dahms pathways. This oxidative pathway is mainly carried out by bacteria.
Figure 1. Three pathways of xylose metabolization by microorganisms: solid light blue, Weinberg pathway; dashed dark blue, Dahms pathway; dashed green, oxidoreductase pathway; and solid red, isomerase pathway. The enzymes are abbreviated as follows: acetyl-CoA synthase (ACS); acetate kinase (AK); enolase (EL); glyceraldehyde-3-phosphate dehydrogenase (GPDH); 2-keto-3-deoxyxylonate dehydratase (KDY); α-ketoglutarate semialdehyde dehydrogenase (KSH); pyrophosphate-acetate phosphotransferase (PAP); pyruvate dehydrogenase complex (PDC); 3-phosphoglycerate kinase (PGK); phosphoketolase (PK); phosphoketolase (PKL); xylonate dehydratase (XDY); xylose dehydrogenase (XDH); xylitol dehydrogenase (XOHDH); xylose isomerase (XI); xylulokinase (XK); xylonolactonase (XLS); and xylose reductase (XR). Adapted from [30][31].
Regarding the isomerase pathway, xylose is first isomerized to xylulose by the enzyme xylose isomerase (
Three pathways of xylose metabolization by microorganisms: solid light blue, Weinberg pathway; dashed dark blue, Dahms pathway; dashed green, oxidoreductase pathway; and solid red, isomerase pathway. The enzymes are abbreviated as follows: acetyl-CoA synthase (ACS); acetate kinase (AK); enolase (EL); glyceraldehyde-3-phosphate dehydrogenase (GPDH); 2-keto-3-deoxyxylonate dehydratase (KDY); α-ketoglutarate semialdehyde dehydrogenase (KSH); pyrophosphate-acetate phosphotransferase (PAP); pyruvate dehydrogenase complex (PDC); 3-phosphoglycerate kinase (PGK); phosphoketolase (PK); phosphoketolase (PKL); xylonate dehydratase (XDY); xylose dehydrogenase (XDH); xylitol dehydrogenase (XOHDH); xylose isomerase (XI); xylulokinase (XK); xylonolactonase (XLS); and xylose reductase (XR). Adapted from [79,80].
Regarding the isomerase pathway, xylose is first isomerized to xylulose by the enzyme xylose isomerase (
). Xylulose is then phosphorylated to xylulose-5-phosphate by XK (
Figure 1). Genes coding for both enzymes are induced by xylose and repressed by glucose and other more readily usable substrates, according to the CCR mechanism, previously reported [25][32]. Xylose isomerase may be activated in the presence of divalent ions such as Mg ). Genes coding for both enzymes are induced by xylose and repressed by glucose and other more readily usable substrates, according to the CCR mechanism, previously reported [62,81]. Xylose isomerase may be activated in the presence of divalent ions such as Mg , whereas others such as Ca
2+ act as inhibitors [6]. This enzyme may also be strongly inhibited by polyol xylitol, especially at a pH below 6 [33][34].
The metabolism of xylulose-5-phosphate continues generally via the PPP pathway, entering the central carbon metabolism. The PPP consists of several reversible transketolase and transaldolase reactions, of which the main function is to produce NAD(P)H, thus providing the reducing power for biosynthetic reactions [35]. Some bacteria, however (e.g., Clostridium acetobutylicum, Clostridium beijerinckii, and Lactococcus lactis), present an alternative mechanism, cleaving xylulose-5-P into acetylphosphate and glyceraldehyde-3-phosphate in a reaction catalyzed by phosphoketolase [36][37][38].
The oxidoreductase pathway is more common in yeast and fungi and uses two steps for the conversion of xylose in xylulose, employing the enzymes XR and XDH ( act as inhibitors [41]. This enzyme may also be strongly inhibited by polyol xylitol, especially at a pH below 6 [82,83].
The metabolism of xylulose-5-phosphate continues generally via the PPP pathway, entering the central carbon metabolism. The PPP consists of several reversible transketolase and transaldolase reactions, of which the main function is to produce NAD(P)H, thus providing the reducing power for biosynthetic reactions [84]. Some bacteria, however (e.g., Clostridium acetobutylicum, Clostridium beijerinckii, and Lactococcus lactis), present an alternative mechanism, cleaving xylulose-5-P into acetylphosphate and glyceraldehyde-3-phosphate in a reaction catalyzed by phosphoketolase [85,86,87].
The oxidoreductase pathway is more common in yeast and fungi and uses two steps for the conversion of xylose in xylulose, employing the enzymes XR and XDH (
Figure 1), with xylitol being an intermediate in this conversion [39]. Xylulose is then further metabolized via XK and PPP ( ), with xylitol being an intermediate in this conversion [88]. Xylulose is then further metabolized via XK and PPP ( ). Either via the isomerase or the oxidoredutase pathways, carbon exits the sugar-phosphate pool by various routes, with one of them being responsible for the formation of pyruvate. The pyruvate undergoes an oxidative decarboxylation to form acetyl-coenzyme A (acetyl-CoA), which is further oxidized via the Krebs cycle (KC) to generate energy.
In the non-phosphorylative pathway, xylose can be converted either to α-ketoglutarate (Weimberg route) or to pyruvate and glucoaldehyde (Dahms route) via a 2-keto-3-deoxy-xylonate intermediate. Burkholderia xenovorans, Caulobacter crescentus, Gluconobacter oxydans, Paraburkholderia sacchari, Pseudomonas fragi, and Pseudomonas taiwanensis are examples of native bacteria that use this pathway to metabolize xylose, with xylose being converted into D-xylonolactone via XDH (
Figure 1) [29][40][41][42]. D-xylonolactone is in turn converted to D-xylonate, which is further converted to 2-keto-3-deoxy-xylonate, the common intermediate to both routes. In the Weimberg pathway (first discovered in P. fragi), 2-keto-3-deoxy-xylonate can be metabolized to α-ketoglutarate semialdehyde and then to α-ketoglutarate, a KC intermediate [41]. Alternatively, in the Dahms route, 2-keto-3-deoxy-xylonate can be cleaved to pyruvate and glucoaldehyde [43]. While pyruvate is then directly converted to acetyl-CoA and enters the central carbon metabolism (CCM), glucoaldehyde is incorporated into CCM at the level of glyoxylate, which requires two reactions generating two reduced equivalents. This has been observed in the hyperthermophilic archaea Sulfolobus solfataricus [44].
The non-phosphorylative pathway has several advantages compared with the isomerase and oxidoredutase pathways because it directly converts D-xylose towards pyruvate and α-ketoglutarate (a KC cycle intermediate) by-passing the PPP, thus minimizing the number of enzymatic steps and the usage of ATP. In addition to saving P, the Weimberg pathway also conserves carbon during xylose metabolism and is thus a very efficient route. The positive aspects of the oxidative pathway promoted the development of recombinant strains using this pathway for the production of different valuable compounds as reviewed by Bañares et al. [45].
A genome analysis of 492 bacteria strains of Bacillus, Bifidobacterium, Caulobacter, Corynebacterium, Escherichia, Lactobacillus, and Xanthomonas and the search for genes involved in xylose metabolism have been performed. The analysis revealed that the pathways of XI and Weimberg were the prevalent routes, with the XI pathway being the best preserved ( ) [78,89,90,91]. D-xylonolactone is in turn converted to D-xylonate, which is further converted to 2-keto-3-deoxy-xylonate, the common intermediate to both routes. In the Weimberg pathway (first discovered in P. fragi), 2-keto-3-deoxy-xylonate can be metabolized to α-ketoglutarate semialdehyde and then to α-ketoglutarate, a KC intermediate [90]. Alternatively, in the Dahms route, 2-keto-3-deoxy-xylonate can be cleaved to pyruvate and glucoaldehyde [92]. While pyruvate is then directly converted to acetyl-CoA and enters the central carbon metabolism (CCM), glucoaldehyde is incorporated into CCM at the level of glyoxylate, which requires two reactions generating two reduced equivalents. This has been observed in the hyperthermophilic archaea Sulfolobus solfataricus [93].
The non-phosphorylative pathway has several advantages compared with the isomerase and oxidoredutase pathways because it directly converts D-xylose towards pyruvate and α-ketoglutarate (a KC cycle intermediate) by-passing the PPP, thus minimizing the number of enzymatic steps and the usage of ATP. In addition to saving P, the Weimberg pathway also conserves carbon during xylose metabolism and is thus a very efficient route. The positive aspects of the oxidative pathway promoted the development of recombinant strains using this pathway for the production of different valuable compounds as reviewed by Bañares et al. [97].
A genome analysis of 492 bacteria strains of Bacillus, Bifidobacterium, Caulobacter, Corynebacterium, Escherichia, Lactobacillus, and Xanthomonas and the search for genes involved in xylose metabolism have been performed. The analysis revealed that the pathways of XI and Weimberg were the prevalent routes, with the XI pathway being the best preserved (
Figure 1) [46].
2.3. Metabolic Pathways to Xylitol and Xylonic Acid
2.3.1. Xylitol
) [100].
3.3. Metabolic Pathways to Xylitol and Xylonic Acid
3.3.1. Xylitol
Table 1 summarizes the contents of this section regarding a selection of bacterial strains able to convert xylose into xylitol.
Table 1. List of bacteria (wild type and genetically modified) with potential for D-xylitol production from lignocellulosic biomass (n.a.—information not available).
List of bacteria (wild type and genetically modified) with potential for D-xylitol production from lignocellulosic biomass (n.a.—information not available).
| Corynebacterium glutamicum ATCC13032 |
| xylose |
Yes |
Shake flask |
50.7 |
0.76 |
| ATCC13032 |
xylose |
| 0.42 |
| Yes |
| [ | 55 | ] |
| Corynebacterium glutamicum |
Shake flask |
50.7 |
0.76 |
0.42 |
[136] |
| Corynebacterium glutamicum ATCC31831 |
| |
| rice straw hydrolysate after dilute sulfuric acid pretreatment |
Yes |
| ATCC31831 |
| Shake flask |
42.9 |
| rice straw hydrolysate after dilute sulfuric acid pretreatment |
Yes |
Shake flask |
42.9 |
| 1.1 |
0.37 |
[ | 55 | ] |
| Corynebacterium glutamicum |
1.1 |
0.37 |
[136] |
| xylose |
56.3 |
0.84 |
0.47 |
Escherichia coli
BL21 |
xylose |
Yes |
Shake flask |
9.1 |
1.10 |
0.45 |
[56] |
Escherichia coli
BL21 |
xylose |
Yes |
Shake flask |
9.1 |
1.10 |
0.45 |
[141] |
| 2 L Batch STR |
6.9 |
0.89 |
0.11 |
Escherichia coli
W3110 |
xylose and glucose |
[ | 57 | ] |
Escherichia coli
W3110 |
xylose and glucose |
| Yes |
Shake flask |
5.1 |
0.51 |
| Yes |
| 0.084 |
| Shake flask |
5.1 |
0.51 |
0.084 |
[134] |
| 5 L Fed-batch STR |
39.2 |
0.98 |
1.09 |
Escherichia coli
BL21 |
xylose and glycerol |
Yes |
5 L Fed-batch STR |
| BL21 |
| 27.3 |
n.a. |
1.8 |
| xylose and glycerol |
Yes |
5 L Fed-batch STR |
| [ | 58 | ] |
Escherichia coli
|
27.3 |
n.a. |
1.8 |
[135] |
Gluconobacter
oxydans
ATCC 621 |
xylose |
No |
] |
Gluconobacter
oxydans
ATCC 621 |
xylose |
| 3 L Batch STR |
109 |
0.95 |
2.5 |
| No |
3 L Batch STR |
| [ |
| 109 |
0.95 |
| 59 |
| 2.5 |
[ | 127 | ] |
| steamed and enzymatically hydrolyzed birchwood |
12.4 |
0.50 |
n.a. |
Gluconobacter
oxydans
DSM 2003 |
corn stover hydrolysate after dry dilute acid pretreatment |
No |
| Gluconobacter |
| 3 L Batch STR |
38.9 |
oxydans
|
| 0.9 |
| DSM 2003 |
corn stover hydrolysate after dry dilute acid pretreatment |
| n.a. |
| No |
| [ | 60 | ] |
| 3 L Batch STR |
38.9 |
0.9 |
n.a. |
[139] |
Gluconobacter
oxydans
DSM 2003 |
5.5 |
[ | 61 | ] |
Gluconobacter
oxydans |
| xylose |
|
| No |
3 L Batch STR |
| DSM 2003 |
| 66.4 |
n.a. |
| xylose |
No |
3 L Batch STR |
66.4 |
n.a. |
5.5 |
[142] |
Gluconobacter
oxydans NL71 |
xylose |
No |
Compressed oxygen-supplied sealed stirred tank reactor (COS-SSTR); pure oxygen supply |
586.3 |
0.95 |
4.7 |
[62] |
Gluconobacter
oxydans NL71 |
xylose |
No |
Compressed oxygen-supplied sealed stirred tank reactor (COS-SSTR); pure oxygen supply |
586.3 |
0.95 |
4.7 |
[138] |
| corn stover diluted sulfuric acid hydrolysates without detoxification |
143.9 |
0.97 |
1.0 |
Gluconobacter
oxydans NL71 |
xylose in distillation stillage of cellulosic ethanol
fermentation broth |
No |
COS-SSTR; fed-batch addition of xylose with cell-recycling |
1813 g in 6-fold cell recycling; 1 L culture medium |
n.a. |
16.8 g h |
| −1 | in 108 h |
[ | 63 | ] |
| in 108 h |
[ | 140 | ] |
Gluconobacter
oxydans NL71 |
corn stover hydrolysate after dry diluted acid pretreatment |
No |
Two-stage fermentation in a 3 L COS-SSTR bioreactor with cell recycling |
167.4 g from 1 kg corn stover |
0.97 |
oxydans NL71 |
| 3.7 |
| corn stover hydrolysate after dry diluted acid pretreatment |
| [ | 64 | ] |
| Gluconobacter |
No |
Two-stage fermentation in a 3 L COS-SSTR bioreactor with cell recycling |
167.4 g from 1 kg corn stover |
0.97 |
3.7 |
[143] |
Gluconobacter
oxydans
ATCC 621 |
xylose |
No |
Fed-batch bioreactor; Immobilized whole-cells; pressurized pure oxygen supply followed by electrodialysis acid chamber (POA-SSB-OE) |
329.2 g xylonic acid |
n.a. |
7.1 g h |
| −1 | in 48 h |
[ | 65 | ] |
| in 48 h |
[ | 144 | ] |
Klebsiella
pneumoniae
(modified) |
bamboo hydrolysate |
|
| Yes |
Fed-batch cultivations |
| (modified) |
| 103 |
0.98 |
| bamboo hydrolysate |
Yes |
Fed-batch cultivations |
103 |
| n.a. |
[ | 66 | ] |
Klebsiella
pneumoniae |
0.98 |
n.a. |
[129] |
Paraburkholderia sacchari
DSM 17165 |
xylose |
No |
2 L Fed-batch STR
xylose as carbon source; high dissolved oxygen concentration |
31] |
Paraburkholderia sacchari
DSM 17165 |
| 150 |
0.85 |
1.5 |
| xylose |
No |
2 L Fed-batch STR
xylose as carbon source; high dissolved oxygen concentration |
| [ |
| 150 |
0.85 |
1.5 |
[80] |
| xylose and glucose |
2 L Fed-batch STR
high dissolved oxygen concentration |
390 |
1.1 |
6.0 |
| Strains |
C-Source |
Genetic Modification |
Growth Conditions |
Xylitol
(g L−1) * |
Yxylitol/xylose
(g g−1) |
Productivity
(g L−1 h−1) * |
Ref. |
|---|
Corynebacterium glutamicum
Cg-ax3 |
arabinose
glucose
xylose |
Yes |
Batch shake flask |
6.7 |
n.a. |
n.a. |
[122] |
| Fed-batch shake flask |
31 |
n.a. |
0.28 gg−1cdw h−1 |
| acid pre-treated liquor of sorghum stover |
Fed-batch shake flask |
27 |
n.a. |
0.22 gg−1cdw h−1 |
| Corynebacterium sp. NRRL B 4247 |
xylose |
No |
Shake flask |
1.7 |
0.57 |
0.071 |
[108] |
6-phosphogluconate (source of NADPH) added to the medium
Shake flask |
10 |
n.a. |
0.067 |
| Corynebacterium sp. no. 208 |
xylose |
No |
6-phosphogluconate (source of NADPH) was added to the medium
Shake flask |
69 |
n.a. |
0.21 |
[123] |
Enterobacter
liquefaciens 553 |
xylose |
No |
Shake flask |
33 |
n.a. |
0.35 |
[110] |
| E. coli BL21(DE3) |
xylose |
Yes |
Shake flask |
202 |
1.0 |
6.37 |
[113] |
| Escherichia coli IS5-d |
xylose and glucose |
Yes |
5 L Batch STR |
110 |
n.a. |
3.06 |
[112] |
| Escherichia coli IS5-M |
corncob hemicellulosic hydrolysate and 24 g L−1 corn steep liquor |
Yes |
15 L Fed-batch STR |
144 |
n.a. |
1.84 |
[112] |
| Escherichia coli HK402 |
xylose and glucose |
Yes |
15 L Fed-batch STR |
172 |
>0.95 |
1.57 |
[112] |
| detoxified hemicellulosic hydrolysate and glucose |
150 |
>0.95 |
1.40 |
| Escherichia coli WZ51 |
detoxified hemicellulosic hydrolysate |
Yes |
15 L Fed-batch STR |
132 |
0.95 |
2.09 |
[114] |
| Mycobacterium smegmatis |
xylose |
No |
immobilized D-xylose isomerase from Bacillus coagulans and immobilized M. smegmatis
Shake flask |
5 g |
0.80 |
n.a. |
[106] |
Paraburkholderia sacchari
DSM 17165 |
xylose |
No |
2 L Fed-batch STR |
17 |
n.a. |
0.39 |
[60] |
Paraburkholderia sacchari
DSM 17165 |
xylose |
No |
2 L Fed-batch STR |
70 |
0.39 |
0.50 |
[80] |
| C-Source |
Genetic Modification |
Growth Conditions |
Xylitol
(g L−1) * |
Yxylitol/xylose
(g g−1) |
Productivity
(g L−1 h−1) * |
Ref. |
|---|
Corynebacterium glutamicum
Cg-ax3 |
arabinose
glucose
xylose |
Yes |
Batch shake flask |
6.7 |
n.a. |
n.a. |
[47] |
| Fed-batch shake flask |
31 |
n.a. |
0.28 gg−1cdw h−1 |
| acid pre-treated liquor of sorghum stover |
Fed-batch shake flask |
27 |
n.a. |
0.22 gg−1cdw h−1 |
| Corynebacterium sp. NRRL B 4247 |
xylose |
No |
Shake flask |
1.7 |
0.57 |
0.071 |
[48] |
6-phosphogluconate (source of NADPH) added to the medium
Shake flask |
10 |
n.a. |
0.067 |
| Corynebacterium sp. no. 208 |
xylose |
No |
6-phosphogluconate (source of NADPH) was added to the medium
Shake flask |
69 |
n.a. |
0.21 |
[49] |
Enterobacter
liquefaciens 553 |
xylose |
No |
Shake flask |
33 |
n.a. |
0.35 |
[50] |
| E. coli BL21(DE3) |
xylose |
Yes |
Shake flask |
202 |
1.0 |
6.37 |
[51] |
| Escherichia coli IS5-d |
xylose and glucose |
Yes |
5 L Batch STR |
110 |
n.a. |
3.06 |
[52] |
| Escherichia coli IS5-M |
corncob hemicellulosic hydrolysate and 24 g L−1 corn steep liquor |
Yes |
15 L Fed-batch STR |
144 |
n.a. |
1.84 |
[52] |
| Escherichia coli HK402 |
xylose and glucose |
Yes |
15 L Fed-batch STR |
172 |
>0.95 |
1.57 |
[52] |
| detoxified hemicellulosic hydrolysate and glucose |
150 |
>0.95 |
1.40 |
| Escherichia coli WZ51 |
detoxified hemicellulosic hydrolysate |
Yes |
15 L Fed-batch STR |
132 |
0.95 |
2.09 |
[53] |
| Mycobacterium smegmatis |
xylose |
No |
immobilized D-xylose isomerase from Bacillus coagulans and immobilized M. smegmatis
Shake flask |
5 g |
0.80 |
n.a. |
[54] |
Paraburkholderia sacchari
DSM 17165 |
xylose |
No |
2 L Fed-batch STR |
17 |
n.a. |
0.39 |
[23] |
Paraburkholderia sacchari
DSM 17165 |
xylose |
No |
2 L Fed-batch STR |
70 |
0.39 |
0.50 |
[31] |
* unless otherwise indicated.
2.3.2. Xylonic Acid
Table 2 gathers information on various examples of bacterial strains able to convert xylose into xylonic acid.
3. Conclusions
Lignocellulosic waste is abundant and renewable; hence, biological processes using xylose-rich lignocellulosic hydrolysates as a carbon source can dramatically reduce the share of raw material-related costs while helping to achieve process sustainability. The hydrolysis of the hemicellulose of lignocellulosic biomass gives rise to a mixture of oligo- and monosaccharides, of which xylose is usually the main component. Although hexoses are generally preferred by most microorganisms, xylose can also be metabolized into commercially relevant molecules by several microorganisms.