
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mehdi Mirsaeidi | + 1993 word(s) | 1993 | 2021-08-16 07:52:49 | | | |
| 2 | Felix Wu | Meta information modification | 1993 | 2021-08-17 02:52:21 | | |
Video Upload Options
Nucleotide-binding oligomerization domain 2 (NOD2) is a cytoplasmic receptor that recognizes invading molecules and danger signals inside the cells.
1. Introduction
2. NLR Family and Structure
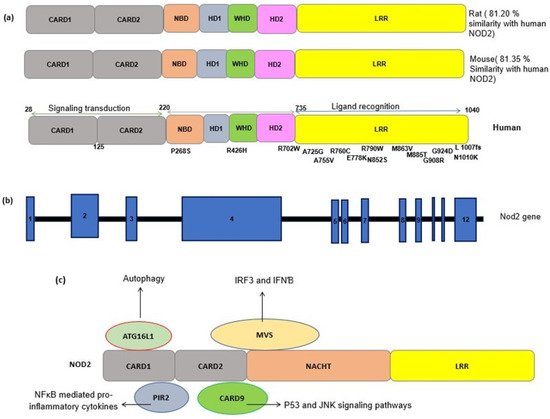
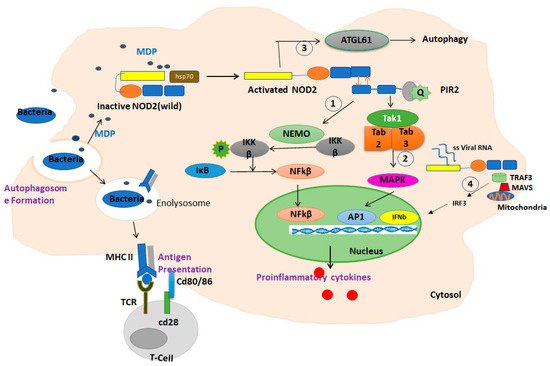
3. NOD2: Cellular and Molecular Mechanisms
4. NOD2 Genetics and Polymorphism
|
Number |
SNPs |
Mutation |
Location |
Population |
Result |
Infection (Disease) |
Method |
Ref |
|---|---|---|---|---|---|---|---|---|
|
1 |
P268S |
CCC > TCC |
NBD domain |
African Americans |
Minor allele T is associated with a decreased risk of TB (Protective) |
Tuberculosis |
Sequencing of the coding regions of the NOD2 gene |
[24] |
|
R702W |
CGG > TGG [14]4 |
HD2 Exon 4 |
Minor allele T is associated with a decreased risk of TB(Protective) |
|||||
|
A725G |
GCT > GGT |
HD2 Exon 4 |
the minor allele G increased the risk of TB |
|||||
|
2 |
R702W |
CGG > TGG |
South African |
No association |
Inflammatory bowel disease (CD & UC) |
PCR of the Exons 4, 8 and 11- HEX-SSCP &RFLP |
[25] |
|
|
A725G |
GCT > GGT |
Increased risk of TB |
||||||
|
G908R |
Rs2066845 |
No association |
||||||
|
1007fs(insC3020) |
L1007P rs5743293 |
No association |
||||||
|
3 |
rs3135499 |
Promoter |
Han Chinese from Jiangsu Province |
T genotype protective |
Tuberculosis |
TaqMan-based allelic discrimination system |
[26] |
|
|
rs7194886 |
Promoter |
Increased risk for T allele carriers |
||||||
|
rs8057341 |
Promoter |
|||||||
|
rs9302752 |
Promoter |
T genotype protective |
||||||
|
4 |
insC3020 |
rs5743293 |
Sardinian population. |
Significant Association (Increased the susceptibility) |
CD & Mycobacterium avium subsp. paratuberculosis |
PCR & sequencing |
[27] |
|
|
R702W |
Rs2066844 |
|||||||
|
G908R |
Rs2066845 |
|||||||
|
5 |
insC3020 |
1007fs |
northern Indian states |
No mutation was observed in the patients and controls |
TB and leprosy |
PCR-RFLP confirmed by gene sequencing |
[28] |
|
|
R702W |
Rs2066844 |
|||||||
|
G908R |
rs2066845 |
|||||||
|
6 |
R702W |
South African |
No association |
Tuberculosis |
Tag Man platform genotyping |
[29] |
||
|
G908R |
||||||||
|
insC3020 |
||||||||
|
7 |
P268S |
C > T rs2066842 |
Exon 4 |
Caucasian patients |
No association |
Sarcoidosis |
Tag Man platform genotyping |
[30] |
|
R587R |
T > G rs1861759 |
Exon 4 |
||||||
|
R702W |
C > T rs2066844 |
Exon 4 |
||||||
|
G908R |
G > C rs2066845 |
Exon 8 |
||||||
|
insC3020 |
rs2066847 |
Exon 11 |
||||||
|
8 |
P268S |
Turkish population |
Association with CD |
Crohn’s Disease and Ulcerative Colitis |
PCR-RFLP |
[31] |
||
|
M863V |
No mutant was found |
|||||||
|
9 |
R702W |
rs2066844 CGG > TGG |
Meta analysis |
C allele is a risk factor |
sarcoidosis |
Meta-analysis |
[32] |
|
|
G908R |
rs2066845 |
no associated |
||||||
|
insC3020 |
rs2066847 |
no associated |
||||||
|
R587R |
rs1861759 |
no associated |
||||||
|
10 |
C-159 T |
rs2569190 |
Meta analysis |
GG is common in TB |
Tuberculosis |
Meta-analysis |
[33] |
|
|
A-1145G |
rs2569191 |
T allele is a risk factor in TB |
||||||
|
IV |
rs1861759 |
TG genotype is higher in TB |
||||||
|
rs7194886 |
T allele is a risk factor of TB |
|||||||
|
R702W |
rs2066844 |
CC genotype is a risk factor for TB |
||||||
|
P 507 T/S |
rs2066842 C > A/T |
CC genotype is a risk factor for TB |
||||||
|
11 |
-159C > T |
-159C > T |
promoter of CD14 |
Chinese |
Higher risk increased promoter activity/increased sNOD2 |
spinal TB |
Seq. |
[34] |
|
12 |
G-1619A |
rs2915863 |
promoter of CD14 |
Han Chinese |
Increased susceptibly/ increased sNOD2 |
tuberculosis |
PCR and seq |
[35] |
|
T-1359G |
rs3138078 |
|||||||
|
A-1145G |
rs2569191 |
|||||||
|
C-159T |
rs2569190 |
|||||||
|
13 |
C(-159)T |
promoter of CD14 |
Han Chinese |
T allele is a RF |
tuberculosis |
PCR-DNA sequencing |
[36] |
|
|
G(-1145)A |
G allele is a RF |
|||||||
|
14 |
C(-159)T |
promoter of CD14 |
increased level of serum soluble CD14 |
tuberculosis |
[37] |
|||
|
15 |
C(-159)T |
promoter of CD14 |
Mexico |
increased Tb susceptibility/ increased level of serum soluble CD14 |
PCR-RFLP |
[38] |
||
|
16 |
C(-159)T |
Promoter |
Meta analysis |
increased risk of TB |
Meta-analysis |
[39] |
||
|
17 |
R426H |
rs562225614 G > A |
Exon 4 |
Case report |
Early Onset Inflammatory Bowel Phenotype |
IBD-Increased expression of inflammatory cytokines |
Sequencing |
[40] |
|
18 |
N1010K |
3030A > C |
LRR domain Exon 12 |
CD |
Sequencing |
[41] |
References
- Carrillo, J.L.M.; García, F.P.C.; Coronado, O.G.; García, M.A.M.; Cordero, J.F.C. Physiology and Pathology of Innate Immune Response against Pathogens; IntechOpen: London, UK, 2017.
- Negroni, A.; Pierdomenico, M.; Cucchiara, S.; Stronati, L. NOD2 and inflammation: Current insights. J. Inflamm. Res. 2018, 11, 49.
- Proell, M.; Riedl, S.J.; Fritz, J.H.; Rojas, A.M.; Schwarzenbacher, R. The Nod-like receptor (NLR) family: A tale of similarities and differences. PLoS ONE 2008, 3, e2119.
- Dolasia, K.; Bisht, M.K.; Pradhan, G.; Udgata, A.; Mukhopadhyay, S. TLRs/NLRs: Shaping the landscape of host immunity. Int. Rev. Immunol. 2018, 37, 3–19.
- Ye, Z.; Ting, J.P. NLR, the nucleotide-binding domain leucine-rich repeat containing gene family. Curr. Opin. Immunol. 2008, 20, 3–9.
- King, A.E.; Horne, A.W.; Hombach-Klonisch, S.; Mason, J.; Critchley, H.O. Differential expression and regulation of nuclear oligomerization domain proteins NOD1 and NOD2 in human endometrium: A potential role in innate immune protection and menstruation. Mol. Hum. Reprod. 2009, 15, 311–319.
- Strober, W.; Murray, P.J.; Kitani, A.; Watanabe, T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat. Rev. Immunol. 2006, 6, 9–20.
- Roth, S.A.; Simanski, M.; Rademacher, F.; Schroder, L.; Harder, J. The pattern recognition receptor NOD2 mediates Staphylococcus aureus-induced IL-17C expression in keratinocytes. J. Investig. Dermatol. 2014, 134, 374–380.
- Trindade, B.C.; Chen, G.Y. NOD1 and NOD2 in inflammatory and infectious diseases. Immunol. Rev. 2020, 297, 139–161.
- Motta, V.; Soares, F.; Sun, T.; Philpott, D.J. NOD-like receptors: Versatile cytosolic sentinels. Physiol. Rev. 2015, 95, 149–178.
- Kersse, K.; Bertrand, M.J.; Lamkanfi, M.; Vandenabeele, P. NOD-like receptors and the innate immune system: Coping with danger, damage and death. Cytokine Growth Factor Rev. 2011, 22, 257–276.
- Franchi, L.; Park, J.H.; Shaw, M.H.; Marina-Garcia, N.; Chen, G.; Kim, Y.G.; Núñez, G. Intracellular NOD-like receptors in innate immunity, infection and disease. Cell. Microbiol. 2008, 10, 1–8.
- Strober, W.; Watanabe, T. NOD2, an intracellular innate immune sensor involved in host defense and Crohn’s disease. Mucosal Immunol. 2011, 4, 484–495.
- Morgulis, A.; Coulouris, G.; Raytselis, Y.; Madden, T.L.; Agarwala, R.; Schäffer, A.A. Database indexing for production MegaBLAST searches. Bioinformatics 2008, 24, 1757–1764.
- Jacob, F.; Vernaldi, S.; Maekawa, T. Evolution and conservation of plant NLR functions. Front. Immunol. 2013, 4, 297.
- Caruso, R.; Warner, N.; Inohara, N.; Nunez, G. NOD1 and NOD2: Signaling, host defense, and inflammatory disease. Immunity 2014, 41, 898–908.
- Shanahan, M.T.; Carroll, I.M.; Grossniklaus, E.; White, A.; von Furstenberg, R.J.; Barner, R.; Fodor, A.A.; Henning, S.J.; Sartor, R.B.; Gulati, A.S. Mouse Paneth cell antimicrobial function is independent of Nod2. Gut 2014, 63, 903–910.
- Homer, C.R.; Richmond, A.L.; Rebert, N.A.; Achkar, J.P.; McDonald, C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology 2010, 139, 1630–1641.
- Brooks, M.N.; Rajaram, M.V.; Azad, A.K.; Amer, A.O.; Valdivia-Arenas, M.A.; Park, J.H.; Nunez, G.; Schlesinger, L.S. NOD2 controls the nature of the inflammatory response and subsequent fate of Mycobacterium tuberculosis and M. bovis BCG in human macrophages. Cell. Microbiol. 2011, 13, 402–418.
- Dominguez-Martinez, D.A.; Nunez-Avellaneda, D.; Castanon-Sanchez, C.A.; Salazar, M.I. NOD2: Activation during bacterial and viral infections, polymorphisms and potential as therapeutic target. Rev. Investig. Clin. 2018, 70, 18–28.
- Negroni, A.; Colantoni, E.; Vitali, R.; Palone, F.; Pierdomenico, M.; Costanzo, M.; Cesi, V.; Cucchiara, S.; Stronati, L. NOD2 induces autophagy to control AIEC bacteria infectiveness in intestinal epithelial cells. Inflamm. Res. 2016, 65, 803–813.
- Beynon, V.; Cotofana, S.; Brand, S.; Lohse, P.; Mair, A.; Wagner, S.; Mussack, T.; Ochsenkühn, T.; Folwaczny, M.; Folwaczny, C. NOD2/CARD15 genotype influences MDP-induced cytokine release and basal IL-12p40 levels in primary isolated peripheral blood monocytes. Inflamm. Bowel Dis. 2008, 14, 1033–1040.
- Mukherjee, T.; Hovingh, E.S.; Foerster, E.G.; Abdel-Nour, M.; Philpott, D.J.; Girardin, S.E. NOD1 and NOD2 in inflammation, immunity and disease. Arch. Biochem. Biophys. 2019, 670, 69–81.
- Austin, C.M.; Ma, X.; Graviss, E.A. Common nonsynonymous polymorphisms in the NOD2 gene are associated with resistance or susceptibility to tuberculosis disease in African Americans. J. Infect. Dis. 2008, 197, 1713–1716.
- Zaahl, M.; Winter, T.; Warnich, L.; Kotze, M. Analysis of the three common mutations in the CARD15 gene (R702W, G908R and 1007fs) in South African colored patients with inflammatory bowel disease. Mol. Cell. Probes. 2005, 19, 278–281.
- Pan, H.; Dai, Y.; Tang, S.; Wang, J. Polymorphisms of NOD2 and the risk of tuberculosis: A validation study in the Chinese population. Int. J. Immunogenet. 2012, 39, 233–240.
- Sechi, L.A.; Gazouli, M.; Ikonomopoulos, J.; Lukas, J.C.; Scanu, A.M.; Ahmed, N.; Fadda, G.; Zanetti, S. Mycobacterium avium subsp. paratuberculosis, genetic susceptibility to Crohn’s disease, and Sardinians: The way ahead. J. Clin. Microbiol. 2005, 43, 5275–5277.
- Singh, V.; Gaur, R.; Mittal, M.; Biswas, S.; Das, R.; Girdhar, B.; Bajaj, B.; Katoch, V.; Kumar, A.; Mohanty, K. Absence of nucleotide-binding oligomerization domain-containing protein 2 variants in patients with leprosy and tuberculosis. Int. J. Immunogenet. 2012, 39, 353–356.
- Möller, M.; Nebel, A.; Kwiatkowski, R.; van Helden, P.D.; Hoal, E.G.; Schreiber, S. Host susceptibility to tuberculosis: CARD15 polymorphisms in a South African population. Mol. Cell. Probes 2007, 21, 148–151.
- Sato, H.; Williams, H.; Spagnolo, P.; Abdallah, A.; Ahmad, T.; Orchard, T.; Copley, S.; Desai, S.; Wells, A.; Du Bois, R. CARD15/NOD2 polymorphisms are associated with severe pulmonary sarcoidosis. Eur. Respir. J. 2010, 35, 324–330.
- Diler, S.B.; Polat, F.; Yaraş, S. The P268S and M863V Polymorphisms of the NOD2/CARD15 gene in Crohn’s disease and ulcerative colitis. Cytol. Genet. 2019, 53, 424–429.
- Chen, X.; Zhou, Z.; Zhang, Y.; Cheng, X.; Guo, X.; Yang, X. NOD2/CARD15 gene polymorphisms and sarcoidosis susceptibility: Review and meta-analysis. Sarcoidosis Vasc. Diffus. Lung Dis. 2018, 35, 115.
- Cubillos-Angulo, J.M.; Fernandes, C.D.; Araújo, D.N.; Carmo, C.A.; Arriaga, M.B.; Andrade, B.B. The influence of single nucleotide polymorphisms of NOD2 or CD14 on susceptibility to tuberculosis: A systematic review. Syst. Rev. 2021, 10, 174.
- Zheng, M.; Shi, S.; Wei, W.; Zheng, Q.; Wang, Y.; Ying, X.; Lu, D. Correlation between MBL2/CD14/TNF-α gene polymorphisms and susceptibility to spinal tuberculosis in Chinese population. Biosci. Rep. 2018, 38.
- Xue, Y.; Zhao, Z.; Chen, F.; Zhang, L.; Li, G.; Ma, K.; Bai, X.; Zuo, Y. Polymorphisms in the promoter of the CD14 gene and their associations with susceptibility to pulmonary tuberculosis. Tissue Antigens 2012, 80, 437–443.
- Zhao, M.; Xue, Y.; Zhao, Z.; Li, F.; Fan, D.; Wei, L.; Sun, X.; Zhang, X.; Wang, X.; Zhang, Y. Association of CD14 G(-1145)A and C(-159)T polymorphisms with reduced risk for tuberculosis in a Chinese Han population. Genet. Mol. Res. 2012, 11, 3425–3431.
- Alavi-Naini, R.; Salimi, S.; Sharifi-Mood, B.; Davoodikia, A.; Moody, B.; Naghavi, A. Association between the CD14 gene C-159T polymorphism and serum soluble CD14 with pulmonary tuberculosis. Int. J. Tuberc. Lung Dis. 2012, 16, 1383–1387.
- Rosas-Taraco, A.G.; Revol, A.; Salinas-Carmona, M.C.; Rendon, A.; Caballero-Olin, G.; Arce-Mendoza, A.Y. CD14 C(-159)T polymorphism is a risk factor for development of pulmonary tuberculosis. J. Infect. Dis. 2007, 196, 1698–1706.
- Miao, R.; Ge, H.; Xu, L.; Xu, F. CD14–159C/T polymorphism contributes to the susceptibility to tuberculosis: Evidence from pooled 1700 cases and 1816 controls. Mol. Biol. Rep. 2014, 41, 3481–3486.
- Girardelli, M.; Loganes, C.; Pin, A.; Stacul, E.; Decleva, E.; Vozzi, D.; Baj, G.; De Giacomo, C.; Tommasini, A.; Bianco, A.M. Novel NOD2 mutation in early-onset inflammatory bowel phenotype. Inflamm. Bowel Dis. 2018, 24, 1204–1212.
- Frade-Proud’Hon-Clerc, S.; Smol, T.; Frenois, F.; Sand, O.; Vaillant, E.; Dhennin, V.; Bonnefond, A.; Froguel, P.; Fumery, M.; Guillon-Dellac, N. A novel rare missense variation of the NOD2 gene: Evidences of implication in Crohn’s disease. Int. J. Mol. Sci. 2019, 20, 835.





