
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Tao Wang | + 1162 word(s) | 1162 | 2021-07-29 04:34:56 | | | |
| 2 | Peter Tang | Meta information modification | 1162 | 2021-08-07 16:11:02 | | |
Video Upload Options
Manganese peroxidase (MnP) is an oxidoreductase with ligninolytic activity and is a promising biocatalyst for the biodegradation of hazardous environmental contaminants, and especially for dye wastewater decolorization.
1. Introduction
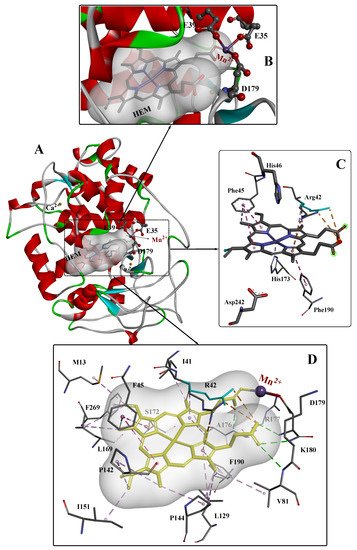
2. The Crystal Structure of MnPs
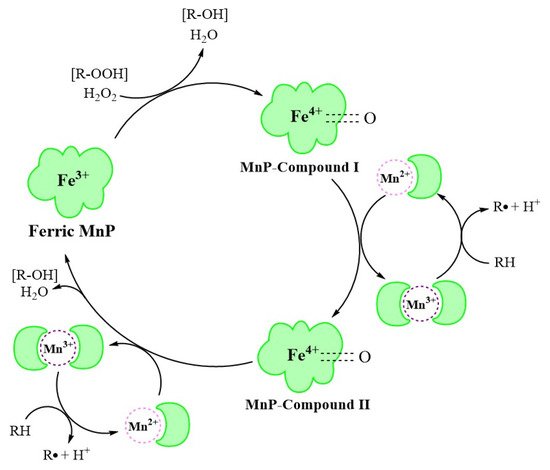
3. MnP Catalysis
4. Application of Unmodified MnPs in the Decolorization of Dye Wastewater
|
Source |
Types of Dyes |
Initial Concentration of Dyes |
Removal Rate |
Time Cost |
Reference |
|---|---|---|---|---|---|
|
Microbial consortium SR |
Crystal Violet |
20 mg/L |
63% |
6 days |
[20] |
|
Cresol Red |
100 mg/L |
93% |
|||
|
CBB G250 |
100 mg/L |
96% |
|||
|
Trametes pubescens strain i8 |
Acid Blue 158 |
50 μM |
95% |
24 h |
[22] |
|
Poly R-478 |
88% |
||||
|
Remazol Brilliant Violet 5R |
76% |
||||
|
Direct Red 5B |
66% |
||||
|
Indigo Carmine |
64% |
||||
|
Methyl Green |
50% |
||||
|
Cibacet Brilliant Blue BG |
46% |
||||
|
Remazol Brilliant Blue Reactif |
42% |
||||
|
Aspergillus terreus GS28 |
Direct Blue-1 |
100 mg/L |
98.4% |
168 h |
[33] |
|
Bjerkandera adusta strain CX-9 |
Acid Blue 158 |
50 μM |
91% |
12 h |
[34] |
|
Poly R-478 |
80% |
||||
|
Cibacet Brilliant Blue BG |
77% |
||||
|
Remazol Brilliant Violet 5R |
70% |
||||
|
Trametes sp.48424 |
Indigo Carmine |
100 mg/L |
94.6% |
18 h |
[35] |
|
Remazol Brilliant Blue R |
85.0% |
||||
|
Remazol Brilliant Violet 5R |
88.4% |
||||
|
Methyl Green |
93.1% |
||||
|
Microbial consortium ZSY |
Metanil Yellow G |
100 mg/L |
93.39% |
48 h |
[36] |
|
Microbial Consortium ZW1 |
Methanil Yellow G |
100 mg/L |
93.3% |
16 h |
[37] |
|
Trichoderma harzianum |
Alizarin Blue Black B |
0.03% |
92.34% |
14 days |
[38] |
|
Phanerochaete chrysosporium CDBB 686 |
Congo Red |
50 ppm |
41.84% |
36 h |
[39] |
|
Poly R-478 |
56.86% |
||||
|
Methyl Green |
69.79% |
||||
|
Bjerkandera adusta CCBAS 930 |
Alizarin Blue Black B |
0.01% |
86.5% |
20 days |
[40] |
|
Acid Blue 129 |
89.22% |
||||
|
Cerrena unicolor BBP6 |
Congo Red |
100 mg/L |
53.9% |
12 h |
[41] |
|
Methyl Orange |
77.6% |
12 h |
|||
|
Remazol Brilliant Blue R |
81.0% |
5 h |
|||
|
Bromophenol Blue |
62.2% |
12 h |
|||
|
Crystal Violet |
80.9% |
12 h |
|||
|
Azure Blue |
63.1% |
24 h |
|||
|
Phanerochaete chrysosporium |
Indigo Carmine |
30 mg/L |
90.18% |
6 h |
[42] |
|
Trametes versicolor |
Dye mixture (Brilliant Blue FCF and Allura Red AC) |
100 mg/L |
80.45% |
14 days |
[43] |
|
Irpex lacteus |
86.04% |
19 days |
|||
|
Bjerkandera adusta |
82.83% |
9 days |
|||
|
Ceriporia lacerata ZJSY |
Congo Red |
100 mg/L |
90% |
48 h |
[44] |
|
Bacillus cohnni RKS9 |
Congo Red |
100 mg/L |
99% |
12 h |
[45] |
|
Schizophyllum commune IBL-06 |
Solar Brilliant Red 80 |
0.01% |
100% |
3 days |
[46] |
|
Irpex lacteus CD2 |
Remazol Brilliant Violet 5R |
50 mg/L |
92.8% |
5 h |
[47] |
|
Remazol Brilliant Blue R |
87.1% |
5 h |
|||
|
Indigo Carmine |
91.5% |
5 h |
|||
|
Direct Red 5B |
82.4% |
36 h |
References
- Verma, A.K.; Dash, R.R.; Bhunia, P. A review on chemical coagulation/flocculation technologies for removal of colour from textile wastewaters. J. Environ. Manag. 2012, 93, 154–168.
- Kishor, R.; Purchase, D.; Saratale, G.D.; Saratale, R.G.; Ferreira, L.F.R.; Bilal, M.; Chandra, R.; Bharagava, R.N. Ecotoxicological and health concerns of persistent coloring pollutants of textile industry wastewater and treatment approaches for environmental safety. J. Environ. Chem. Eng. 2021, 9, 105012.
- Tkaczyk, A.; Mitrowska, K.; Posyniak, A. Synthetic organic dyes as contaminants of the aquatic environment and their implications for ecosystems: A review. Sci. Total. Environ. 2020, 717, 137222.
- Selvaraj, V.; Karthika, T.S.; Mansiya, C.; Alagar, M. An over review on recently developed techniques, mechanisms and intermediate involved in the advanced azo dye degradation for industrial applications. J. Mol. Struct. 2021, 1224, 129195.
- Ihsanullah, I.; Jamal, A.; Ilyas, M.; Zubair, M.; Khan, G.; Atieh, M.A. Bioremediation of dyes: Current status and prospects. J. Water Process. Eng. 2020, 38, 101680.
- Katheresan, V.; Kansedo, J.; Lau, S.Y. Efficiency of various recent wastewater dye removal methods: A review. J. Environ. Chem. Eng. 2018, 6, 4676–4697.
- Ghosh, A.; Dastidar, M.G.; Sreekrishnan, T. Bioremediation of chromium complex dyes and treatment of sludge generated during the process. Int. Biodeterior. Biodegrad. 2017, 119, 448–460.
- Kishor, R.; Saratale, G.D.; Saratale, R.G.; Ferreira, L.F.R.; Bilal, M.; Iqbal, H.M.; Bharagava, R.N. Efficient degradation and detoxification of methylene blue dye by a newly isolated ligninolytic enzyme producing bacterium Bacillus Albus MW407057. Colloids Surf. B Biointerfaces 2021, 206, 111947.
- Gurung, N.; Ray, S.; Bose, S.; Rai, V. A Broader View: Microbial Enzymes and Their Relevance in Industries, Medicine, and Beyond. BioMed Res. Int. 2013, 2013, 329121.
- Vilar, D.D.S.; Bilal, M.; Bharagava, R.N.; Kumar, A.; Nadda, A.K.; Salazar-Banda, G.R.; Eguiluz, K.I.B.; Ferreira, L.F.R. Lignin-modifying enzymes: A green and environmental responsive technology for organic compound degradation. J. Chem. Technol. Biotechnol. 2021, 96, 6751.
- Kuwahara, M.; Glenn, J.K.; Morgan, M.A.; Gold, M.H. Separation and characterization of two extracelluar H2O2-dependent oxidases from ligninolytic cultures of Phanerochaete chrysosporium. FEBS Lett. 1984, 169, 247–250.
- Glenn, J.K.; Gold, M.H. Purification and characterization of an extracellular Mn(II)-dependent peroxidase from the lignin-degrading basidiomycete, Phanerochaete chrysosporium. Arch. Biochem. Biophys. 1985, 242, 329–341.
- Paszczyński, A.; Huynh, V.B.; Crawford, R. Enzymatic activities of an extracellular, manganese-dependent peroxidase from Phanerochaete chrysosporium. FEMS Microbiol. Lett. 1985, 29, 37–41.
- Paszczyński, A.; Huynh, V.B.; Crawford, R. Comparison of ligninase-I and peroxidase-M2 from the white-rot fungus Phanerochaete chrysosporium. Arch. Biochem. Biophys. 1986, 244, 750–765.
- Fernández-Fueyo, E.; Acebes, S.; Ruiz-Dueñas, F.J.; Martínez, M.J.; Romero, A.; Medrano, F.J.; Guallar, V.; Martínez, A.T. Structural implications of the C-terminal tail in the catalytic and stability properties of manganese peroxidases from ligninolytic fungi. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 3253–3265.
- Ruiz-Dueñas, F.J.; Barrasa, J.M.; Sánchez-García, M.; Camarero, S.; Miyauchi, S.; Serrano, A.; Linde, D.; Babiker, R.; Drula, E.; Ayuso-Fernández, I.; et al. Genomic analysis enlightens Agaricales lifestyle evolution and increasing peroxidase diversity. Mol. Biol. Evol. 2021, 38, 1428–1446.
- Ayuso-Fernández, I.; Ruiz-Dueñas, F.J.; Martínez, A.T. Evolutionary convergence in lignin-degrading enzymes. Proc. Natl. Acad. Sci. USA 2018, 115, 6428–6433.
- Singh, A.K.; Bilal, M.; Iqbal, H.M.; Meyer, A.S.; Raj, A. Bioremediation of lignin derivatives and phenolics in wastewater with lignin modifying enzymes: Status, opportunities and challenges. Sci. Total. Environ. 2021, 777, 145988.
- Bilal, M.; Bagheri, A.R.; Vilar, D.S.; Aramesh, N.; Eguiluz, K.I.B.; Ferreira, L.F.R.; Ashraf, S.S.; Iqbal, H.M. Oxidoreductases as a versatile biocatalytic tool to tackle pollutants for clean environment—A review. J. Chem. Technol. Biotechnol. 2021, 96, 1–44.
- Yang, X.; Wang, J.; Zhao, X.; Wang, Q.; Xue, R. Increasing manganese peroxidase production and biodecolorization of triphenylmethane dyes by novel fungal consortium. Bioresour. Technol. 2011, 102, 10535–10541.
- Bilal, M.; Asgher, M. Sandal reactive dyes decolorization and cytotoxicity reduction using manganese peroxidase immobilized onto polyvinyl alcohol-alginate beads. Chem. Cent. J. 2015, 9, 1–14.
- Rekik, H.; Jaouadi, N.Z.; Bouacem, K.; Zenati, B.; Kourdali, S.; Badis, A.; Annane, R.; Bouanane-Darenfed, A.; Bejar, S.; Jaouadi, B. Physical and enzymatic properties of a new manganese peroxidase from the white-rot fungus Trametes pubescens strain i8 for lignin biodegradation and textile-dyes biodecolorization. Int. J. Biol. Macromol. 2019, 125, 514–525.
- Engel, M.; Hoffmann, T.; Wagner, L.; Wermann, M.; Heiser, U.; Kiefersauer, R.; Huber, R.; Bode, W.; Demuth, H.-U.; Brandstetter, H. The crystal structure of dipeptidyl peptidase IV (CD26) reveals its functional regulation and enzymatic mechanism. Proc. Natl. Acad. Sci. USA 2003, 100, 5063–5068.
- Qiu, J.; Wilkens, C.; Barrett, K.; Meyer, A.S. Microbial enzymes catalyzing keratin degradation: Classification, structure, function. Biotechnol. Adv. 2020, 44, 107607.
- Chang, M.; Zhou, Y.; Wang, H.; Liu, Z.; Zhang, Y.; Feng, Y. Crystal structure of the multifunctional SAM-dependent enzyme LepI provides insights into its catalytic mechanism. Biochem. Biophys. Res. Commun. 2019, 515, 255–260.
- Ayuso-Fernández, I.; Martínez, A.T.; Ruiz-Dueñas, F.J. Experimental recreation of the evolution of lignin-degrading enzymes from the Jurassic to date. Biotechnol. Biofuels 2017, 10, 1–13.
- Blodig, W.; Smith, A.; Doyle, W.A.; Piontek, K. Crystal structures of pristine and oxidatively processed lignin peroxidase expressed in Escherichia coli and of the W171F variant that eliminates the redox active tryptophan 171. Implications for the reaction mechanism. J. Mol. Biol. 2001, 305, 851–861.
- Sundaramoorthy, M.; Gold, M.H.; Poulos, T.L. Ultrahigh (0.93A) resolution structure of manganese peroxidase from Phanerochaete chrysosporium: Implications for the catalytic mechanism. J. Inorg. Biochem. 2010, 104, 683–690.
- George, S.J.; Kvaratskhelia, M.; Dilworth, M.J.; Thorneley, R.N.F. Reversible alkaline inactivation of lignin peroxidase involves the release of both the distal and proximal site calcium ions and bishistidine co-ordination of the haem. Biochem. J. 1999, 344, 237–244.
- Hofrichter, M. Review: Lignin conversion by manganese peroxidase (MnP). Enzym. Microb. Technol. 2002, 30, 454–466.
- Wariishi, H.; Huang, J.; Dunford, H.; Gold, M. Reactions of lignin peroxidase compounds I and II with veratryl alcohol. Transient-state kinetic characterization. J. Biol. Chem. 1991, 266, 20694.
- Wariishi, H.; Valli, K.; Gold, M.H.; Wariishi, H.; Valli, K.; Gold, M.H. Manganese(II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium. Kinetic mechanism and role of chelators. J. Biol. Chem. 1992, 267, 23688–23695.
- Singh, G.; Dwivedi, S. Decolorization and degradation of Direct Blue-1 (Azo dye) by newly isolated fungus Aspergillus terreus GS28, from sludge of carpet industry. Environ. Technol. Innov. 2020, 18, 100751.
- Bouacem, K.; Rekik, H.; Jaouadi, N.Z.; Zenati, B.; Kourdali, S.; El Hattab, M.; Badis, A.; Annane, R.; Bejar, S.; Hacene, H.; et al. Purification and characterization of two novel peroxidases from the dye-decolorizing fungus Bjerkandera adusta strain CX-9. Int. J. Biol. Macromol. 2018, 106, 636–646.
- Zhang, H.; Zhang, S.; He, F.; Qin, X.; Zhang, X.; Yang, Y. Characterization of a manganese peroxidase from white-rot fungus Trametes sp.48424 with strong ability of degrading different types of dyes and polycyclic aromatic hydrocarbons. J. Hazard. Mater. 2016, 320, 265–277.
- Guo, G.; Hao, J.; Tian, F.; Liu, C.; Ding, K.; Zhang, C.; Yang, F.; Xu, J. Decolorization of Metanil Yellow G by a halophilic alkalithermophilic bacterial consortium. Bioresour. Technol. 2020, 316, 123923.
- Guo, G.; Hao, J.; Tian, F.; Liu, C.; Ding, K.; Xu, J.; Zhou, W.; Guan, Z. Decolorization and detoxification of azo dye by halo-alkaliphilic bacterial consortium: Systematic investigations of performance, pathway and metagenome. Ecotoxicol. Environ. Saf. 2020, 204, 111073.
- Rybczyńska-Tkaczyk, K.; Święciło, A.; Szychowski, K.; Korniłłowicz-Kowalska, T. Comparative study of eco- and cytotoxicity during biotransformation of anthraquinone dye Alizarin Blue Black B in optimized cultures of microscopic fungi. Ecotoxicol. Environ. Saf. 2018, 147, 776–787.
- Sosa-Martínez, J.D.; Balagurusamy, N.; Montañez, J.; Peralta, R.A.; Moreira, R.D.F.P.M.; Bracht, A.; Peralta, R.M.; Morales-Oyervides, L. Synthetic dyes biodegradation by fungal ligninolytic enzymes: Process optimization, metabolites evaluation and toxicity assessment. J. Hazard. Mater. 2020, 400, 123254.
- Rybczyńska-Tkaczyk, K.; Korniłłowicz-Kowalska, T.; Szychowski, K.; Gmiński, J. Biotransformation and toxicity effect of monoanthraquinone dyes during Bjerkandera adusta CCBAS 930 cultures. Ecotoxicol. Environ. Saf. 2020, 191, 110203.
- Zhang, H.; Zhang, J.; Zhang, X.; Geng, A. Purification and characterization of a novel manganese peroxidase from white-rot fungus Cerrena unicolor BBP6 and its application in dye decolorization and denim bleaching. Process. Biochem. 2018, 66, 222–229.
- Li, H.; Zhang, R.; Tang, L.; Zhang, J.; Mao, Z. Manganese peroxidase production from cassava residue by Phanerochaete chrysosporium in solid state fermentation and its decolorization of indigo carmine. Chin. J. Chem. Eng. 2015, 23, 227–233.
- Merino-Restrepo, A.; Mejía, F.; Velásquez-Quintero, C.; Hormaza-Anaguano, A. Evaluation of several white-rot fungi for the decolorization of a binary mixture of anionic dyes and characterization of the residual biomass as potential organic soil amendment. J. Environ. Manag. 2020, 254, 109805.
- Wang, N.; Chu, Y.; Wu, F.; Zhao, Z.; Xu, X. Decolorization and degradation of Congo red by a newly isolated white rot fungus, Ceriporia lacerata, from decayed mulberry branches. Int. Biodeterior. Biodegrad. 2017, 117, 236–244.
- Kishor, R.; Purchase, D.; Saratale, G.D.; Ferreira, L.F.R.; Bilal, M.; Iqbal, H.M.; Bharagava, R.N. Environment friendly degradation and detoxification of Congo red dye and textile industry wastewater by a newly isolated Bacillus cohnni (RKS9). Environ. Technol. Innov. 2021, 22, 101425.
- Asgher, M.; Yasmeen, Q.; Iqbal, H.M.N. Enhanced decolorization of Solar brilliant red 80 textile dye by an indigenous white rot fungus Schizophyllum commune IBL-06. Saudi J. Biol. Sci. 2013, 20, 347–352.
- Xing, Q.; Zhang, J.; Zhang, X.; Yang, Y. Induction, Purification and Characterization of a Novel Manganese Peroxidase from Irpex lacteus CD2 and Its Application in the Decolorization of Different Types of Dye. PLoS ONE 2014, 9, e113282.





