Manganese peroxidase (MnP) is an oxidoreductase with ligninolytic activity and is a promising biocatalyst for the biodegradation of hazardous environmental contaminants, and especially for dye wastewater decolorization.
1. Introduction
The textile industry produces large quantities of wastewater containing different types of dyes used during the dyeing process, which cause great harm to the environment [
1,
2]. Many dyes and their intermediate metabolites have been identified as mutagenic, teratogenic, or carcinogenic, and represent serious health threats to living ecosystems [
3].
At present, the treatment of dye wastewater mainly relies on physical or chemical management techniques, including chemical reduction, adsorption, ionizing radiation, precipitation, flocculation and flotation, membrane filtration, electric coagulation, electrochemical destruction, and ion exchange ozonation [
4,
5]. These technologies have obvious shortcomings such as the excessive use of chemicals, sludge production, expensive factory requirements or high operating expenses, low decolorization efficiencies, and the inability to handle large numbers of dyes with different structures, so they are not economically suitable for large-scale wastewater decolorization [
6].
The current focus is to reduce toxicity and develop an efficient, economical, and green dye detoxification and decolorization technology. Compared with physical and chemical methods, biological methods offer beneficial and effective prospects due to their economical and environmentally friendly advantages, as well as being simple to use, safe, and efficient, with no secondary pollution [
7,
8]. Therefore, biotechnology is considered the best choice to degrade and remove these pollutants effectively. In the biotechnology field, enzyme biocatalysis is currently the main research area due to its broad application prospects [
9,
10].
Manganese peroxidases (EC 1.11.1.13; MnPs) are a family of heme-containing glycoproteins belonging to the oxidoreductase group. It was discovered in
Phanerochaete chrysosporium and is also found in many bacteria and white-rot fungi (WRF) [
11,
12,
13,
14]. There are different MnPs in nature with differentiated properties. For example, long and short MnPs were reported in WRF associated with the presence/absence of the C-terminal tail extension, and these showed different catalytic and stability properties [
15]. According to the residues of the Mn
2+-binding site, three novel subfamilies of MnP were described in Agaricales including MnP-ESD (Glu/Ser/Asp Mn
2+-oxidation site), MnP-DGD (Asp/Gly/Asp Mn
2+-oxidation site), and MnP-DED (Asp/Glu/Asp Mn
2+-oxidation site) [
16]. However, the Mn
2+-binding site is not the unique feature of MnPs, because versatile peroxidases (VPs), which evolved directly from MnPs, also possess such a site and can oxidize Mn
2+ to Mn
3+ [
17].
For enzyme applications, MnPs can catalyze the peroxide-dependent degradation of a variety of toxic dye pollutants, phenolic compounds, antibiotics, and polycyclic aromatic hydrocarbons, so are promising biocatalysts for hazardous environmental contaminants biodegradation [
18,
19]. Moreover, the use of MnPs is suitable for dye wastewater decolorization as the process is simple and the enzyme can be recycled, thus reducing operating costs [
20,
21,
22].
2. The Crystal Structure of MnPs
The crystal structure of an enzyme provides information on the catalytic mechanism and for potential in-depth design and transformation, and for realizing the green biotechnological use of enzymes [
23,
24,
25].
The heme conformation of MnP is similar to that of lignin peroxidase (LiP) and is evolutionarily conserved [
26]. In its resting-state form, MnP is a strongly helical protein containing a Fe
3+ penta-coordinated structure with the porphyrin ring of the heme cofactor and a proximal histidine, with the sixth coordination position open for H
2O
2 [
27].
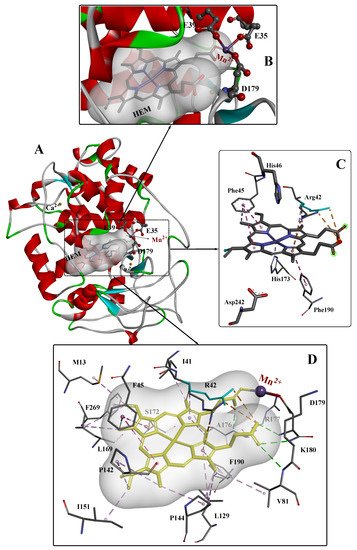
To date, several crystal structures of MnP from different sources have been reported, and the highest-resolution crystal structures (~0.93Å) of MnP complexed with Mn
2+ (Mn-MnP) are shown in
Figure 1 [
28]. The conserved Ca
2+ ions are important for the stability of the protein [
29]; these are indicated as gold yellow spheres and the position of the Mn
2+ substrate is shown in violet. The active site is composed of three highly conserved amino acids (Glu35, Glu39, and Asp179) and one heme propionate. The Mn
2+ substrate binds in the center of the active site, and the heme propionate (HEM) is located in the internal hydrophobic cavity of the enzyme. The spatial structure of HEM is further stabilized by four hydrogen bonds (green dashed line), two electrostatic interactions (orange dashed line), and some other weak interactions. The catalytic site of heme peroxidases is strongly conserved, with only minor variations occurring in the replacement of Phe with Trp in several enzymes such as ascorbate peroxidase and cytochrome c. The Asp–His pair (242 and 173, respectively) is also conserved.
Figure 1. The overall structure (
A), active site structure (
B,
C), and interaction mode (
D) of Mn–MnP refined at 0.93 Å resolution [
28]. PDB ID: 3M5Q.
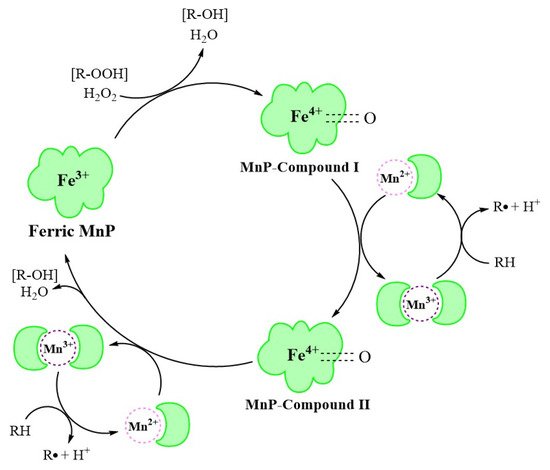
3. MnP Catalysis
At the beginning of the catalytic cycle, H
2O
2 or organic peroxide binds to the enzyme in resting state in ferric (Fe
3+) form (
Figure 2). This process releases one molecule of H
2O and forms MnP–compound I (Fe
4+-oxo-porphyrin radical complex), with two oxidation equivalents. This oxidizes Mn
2+ to Mn
3+, forming MnP–compound II (Fe
4+-oxo-porphyrin complex). Immediately afterwards, the MnP–compound II combines with Mn
2+ in a similar manner to generate Mn
3+, releasing one molecule of H
2O, and is reduced to the original state of ferric MnP, completing the catalytic cycle [
30].
Figure 2. The MnP catalytic cycle [
30].
The MnP catalytic cycle resembles that of other lignin and heme peroxidases in the presence of native Fe
3+ enzymes and two reactive intermediates [
31]. However, in contrast to other peroxidases, MnP preferentially uses Mn
2+ as the substrate, converting it to the strong oxidation state of Mn
3+ through a series of redox reactions [
32].
4. Application of Unmodified MnPs in the Decolorization of Dye Wastewater
Table 1 contains a summary of recent studies on the breakdown and decolorization of textile-derived dye compounds by microbial MnPs.
Table 1. Recent applications of unmodified MnPs in dye decolorization.
|
Source
|
Types of Dyes
|
Initial Concentration of Dyes
|
Removal Rate
|
Time Cost
|
Reference
|
|
Microbial consortium SR
|
Crystal Violet
|
20 mg/L
|
63%
|
6 days
|
[20]
|
|
Cresol Red
|
100 mg/L
|
93%
|
|
CBB G250
|
100 mg/L
|
96%
|
|
Trametes pubescens strain i8
|
Acid Blue 158
|
50 μM
|
95%
|
24 h
|
[22]
|
|
Poly R-478
|
88%
|
|
Remazol Brilliant Violet 5R
|
76%
|
|
Direct Red 5B
|
66%
|
|
Indigo Carmine
|
64%
|
|
Methyl Green
|
50%
|
|
Cibacet Brilliant Blue BG
|
46%
|
|
Remazol Brilliant Blue Reactif
|
42%
|
|
Aspergillus terreus GS28
|
Direct Blue-1
|
100 mg/L
|
98.4%
|
168 h
|
[33]
|
|
Bjerkandera adusta strain CX-9
|
Acid Blue 158
|
50 μM
|
91%
|
12 h
|
[34]
|
|
Poly R-478
|
80%
|
|
Cibacet Brilliant Blue BG
|
77%
|
|
Remazol Brilliant Violet 5R
|
70%
|
|
Trametes sp.48424
|
Indigo Carmine
|
100 mg/L
|
94.6%
|
18 h
|
[36]
|
|
Remazol Brilliant Blue R
|
85.0%
|
|
Remazol Brilliant Violet 5R
|
88.4%
|
|
Methyl Green
|
93.1%
|
|
Microbial consortium ZSY
|
Metanil Yellow G
|
100 mg/L
|
93.39%
|
48 h
|
[37]
|
|
Microbial Consortium ZW1
|
Methanil Yellow G
|
100 mg/L
|
93.3%
|
16 h
|
[38]
|
|
Trichoderma harzianum
|
Alizarin Blue Black B
|
0.03%
|
92.34%
|
14 days
|
[39]
|
|
Phanerochaete chrysosporium CDBB 686
|
Congo Red
|
50 ppm
|
41.84%
|
36 h
|
[40]
|
|
Poly R-478
|
56.86%
|
|
Methyl Green
|
69.79%
|
|
Bjerkandera adusta CCBAS 930
|
Alizarin Blue Black B
|
0.01%
|
86.5%
|
20 days
|
[41]
|
|
Acid Blue 129
|
89.22%
|
|
Cerrena unicolor BBP6
|
Congo Red
|
100 mg/L
|
53.9%
|
12 h
|
[42]
|
|
Methyl Orange
|
77.6%
|
12 h
|
|
Remazol Brilliant Blue R
|
81.0%
|
5 h
|
|
Bromophenol Blue
|
62.2%
|
12 h
|
|
Crystal Violet
|
80.9%
|
12 h
|
|
Azure Blue
|
63.1%
|
24 h
|
|
Phanerochaete chrysosporium
|
Indigo Carmine
|
30 mg/L
|
90.18%
|
6 h
|
[43]
|
|
Trametes versicolor
|
Dye mixture
(Brilliant Blue FCF
and
Allura Red AC)
|
100 mg/L
|
80.45%
|
14 days
|
[44]
|
|
Irpex lacteus
|
86.04%
|
19 days
|
|
Bjerkandera adusta
|
82.83%
|
9 days
|
|
Ceriporia lacerata ZJSY
|
Congo Red
|
100 mg/L
|
90%
|
48 h
|
[45]
|
|
Bacillus cohnni RKS9
|
Congo Red
|
100 mg/L
|
99%
|
12 h
|
[46]
|
|
Schizophyllum commune IBL-06
|
Solar Brilliant Red 80
|
0.01%
|
100%
|
3 days
|
[47]
|
|
Irpex lacteus CD2
|
Remazol Brilliant Violet 5R
|
50 mg/L
|
92.8%
|
5 h
|
[48]
|
|
Remazol Brilliant Blue R
|
87.1%
|
5 h
|
|
Indigo Carmine
|
91.5%
|
5 h
|
|
Direct Red 5B
|
82.4%
|
36 h
|
This entry is adapted from the peer-reviewed paper 10.3390/molecules26154403