
Video Upload Options
The coronavirus disease 2019 (COVID-19) has caused a large global outbreak. It is accordingly important to develop accurate and rapid diagnostic methods. The polymerase chain reaction (PCR)-based method including reverse transcription-polymerase chain reaction (RT-PCR) is the most widely used assay for the detection of SARS-CoV-2 RNA. Along with the RT-PCR method, digital PCR has emerged as a powerful tool to quantify nucleic acid of the virus with high accuracy and sensitivity. Non-PCR based techniques such as reverse transcription loop-mediated isothermal amplification (RT-LAMP) and reverse transcription recombinase polymerase amplification (RT-RPA) are considered to be rapid and simple nucleic acid detection methods and were reviewed in this entry. Non-conventional molecular diagnostic methods including next-generation sequencing (NGS), CRISPR-based assays and nanotechnology are improving the accuracy and sensitivity of COVID-19 diagnosis.
1. Background
1.1. Nomenclature of SARS-CoV-2
| GISAID Clades | PANGO Lineage | Nextstrain Clades | Notable Variants |
|---|---|---|---|
| S | A | 19B | A.23.1 |
| L | B | 19A | Wuhan-Hu-1 |
| V | |||
| G | B.1 | 20A | B.1.525, B.1.627 |
| GH | B.1 | 20C | B.1.427, B.1.429, B.1.526 |
| B.1.2 | 20G | ||
| B.1.596 | |||
| B.1.351 | 20H/501.Y.V2 | B.1.351 | |
| GR | B.1.1.1 | 20B | |
| P.3 | P.3 | ||
| C | 20D | ||
| D.2 | 20F | ||
| P.1 | 20J/501.Y.V3 | P.1 | |
| GV | B.1.177 | 20E (EU1) | B.1.177 |
| GRY | B.1.1.7 | 20I/501.Y.V1 | B.1.1.7 |
1.2. Notable Variants of SARS-CoV-2
| PANGO Lineage | CDC Designation | PHE Designation | First Detected | Spike Protein Substitutions |
|---|---|---|---|---|
| B.1.1.7 | VOC | VOC-20DEC-01, VOC-21FEB-02 * | United Kingdom | 69del, 70del, 144del, (E484K), (S494P), N501Y, A570D, D614G, P681H, T716I, S982A, D1118H (K1191N) |
| B.1.351 | VOC | VOC-20DEC-02 | South Africa | D80A, D215G, 241del, 242del, 243del, K417N, E484K, N501Y, D614G, A701V |
| P.2 | VOI | VUI-21JAN-01 | Brazil | E484K, (F565L), D614G, V1176F |
| P.1 | VOC | VOC-21JAN-02 | Brazil | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I |
| A.23.1 | - | VUI-21FEB-01 * | Uganda | F157L, V367F, (E484K), Q613H, P681R |
| B.1.525 | VOI | VUI-21FEB-03 | United Kingdom | A67V, 69del, 70del, 144del, E484K, D614G, Q677H, F888L |
| B.1.1.318 | - | VUI-21FEB-04 | United Kingdom | D614G, D796H, E484K, P681H, T95I, 144del |
| P.3 | - | VUI-21MAR-02 | Philippines | E484K, N501Y, P681H |
| B.1.617 | VOI | VUI-21APR-01 | India | L452R, E484Q, D614G |
| B.1.617.2 | VOI | VOC-21APR-02 | India | T19R, (G142D), 156del, 157del, R158G, L452R, T478K, D614G, P681R, D950N |
| B.1.617.3 | VOI | VUI-21APR-03 | India | T19R, G142D, L452R, E484Q, D614G, P681R, D950N |
| AV.1 | - | VUI-21MAY-01 | United Kingdom | D80G, T95I, G142D, 144del, N439K, E484K, D614G, P681H, I1130V, D1139H |
| B.1.617.1 | VOI | - | India | (T95I), G142D, E154K, L452R, E484Q, D614G, P681R, Q1071H |
| B.1.526 | VOI | - | United States | (L5F), T95I, D253G, (S477N), (E484K), D614G, (A701V) |
| B.1.526.1 | VOI | - | United States | D80G, 144del, F157S, L452R, D614G, (T791I), (T859N), D950H |
| B.1.427 | VOC | - | United States | L452R, D614G |
| B.1.429 | VOC | - | United States | S13I, W152C, L452R, D614G |
2. PCR-Based SARS-CoV-2 Detection
2.1. Reverse Transcription Quantitative PCR (RT-qPCR) Method
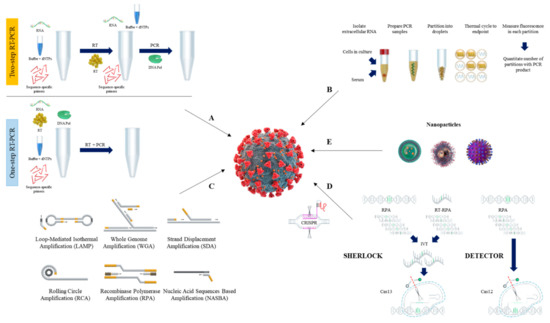
2.2. Reverse Transcription Digital PCR (RT-dPCR) Method
3. Isothermal Nucleic Acid Amplification Methods
3.1. Loop-Mediated Isothermal Amplification (LAMP)
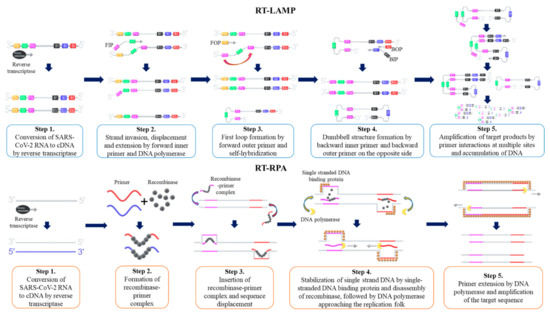
3.2. Recombinase Polymerase Amplification (RPA)
3.3. Other Isothermal Nuleic Acid Amplification Methods
| Method | Components | Temperature | Time | Detection Method | Advantages * | Disadvantages * |
|---|---|---|---|---|---|---|
| Loop-mediated isothermal amplification (LAMP) | DNA polymerase, forward inner primer, backward inner primer, forward outer primer, backward outer primer | 60–65 °C | >1 h | Colorimetric, turbimetric, fluorescence probe, intercalating dye | High specificity. Less sensitive to inhibitors in biological samples | False positive in negative control |
| Recombinase polymerase amplification (RPA) | Recombinase, single stranded binding protein, DNA polymerase, forward primer, reverse primer | 37–42 °C | >1 h | Fluorescence, antigenic-tag (antibody) | Performed in the presence of PCR inhibitors. Fast and sensitive | Inhibited by detergents (SDS and CTAB). Non-specific/high background signal |
| Nucleic acid sequence-based amplification (NASBA) | RNase H, reverse transcriptase, T7 DNA-dependent RNA polymerase, forward primer with T7 promoter sequence, reverse primer | 41 °C | >2 h | Fluorescence | More sensitive and less time-consuming | Non-specific reactions/false positives |
| Strand-displacement amplification (SDA) | DNA polymerase, restriction endonuclease, primers, dCTP, dTTP, dGTP, dATPα | 37–49 °C | >2 h | Fluorescence | High specificity. Detection of large RNA molecules | Non-specific reaction/high background signal |
| Rolling circle amplification (RCA) | DNA ligase, DNA polymerase, primer, padlock probe | 30–37 °C | >1.5 h | Fluorescence | High specificity | False negatives and false positives |
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature 2020, 579, 265–269.
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273.
- Van Der Hoek, L.; Pyrc, K.; Jebbink, M.F.; Vermeulen-Oost, W.; Berkhout, R.J.M.; Wolthers, K.C.; Wertheim-Van Dillen, P.M.E.; Kaandorp, J.; Spaargaren, J.; Berkhout, B. Identification of a New Human Coronavirus. Nat. Med. 2004, 10, 368–373.
- Vabret, A.; Mourez, T.; Gouarin, S.; Petitjean, J.; Freymuth, F. An Outbreak of Coronavirus OC43 Respiratory Infection in Normandy, France. Clin. Infect. Dis. 2003, 36, 985–989.
- Vijgen, L.; Keyaerts, E.; Moës, E.; Thoelen, I.; Wollants, E.; Lemey, P.; Vandamme, A.-M.; Van Ranst, M. Complete Genomic Sequence of Human Coronavirus OC43: Molecular Clock Analysis Suggests a Relatively Recent Zoonotic Coronavirus Transmission Event. J. Virol. 2005, 79, 1595–1604.
- Owczarek, K.; Szczepanski, A.; Milewska, A.; Baster, Z.; Rajfur, Z.; Sarna, M.; Pyrc, K. Early Events during Human Coronavirus OC43 Entry to the Cell. Sci. Rep. 2018, 8, 7124.
- Pene, F.; Merlat, A.; Vabret, A.; Rozenberg, F.; Buzyn, A.; Dreyfus, F.; Cariou, A.; Freymuth, F.; Lebon, P. Coronavirus 229E-Related Pneumonia in Immunocompromised Patients. Clin. Infect. Dis. 2003, 37, 929–932.
- Hendley, J.O.; Fishburne, H.B.; Gwaltney, J.M. Coronavirus Infections in Working Adults. Eight-Year Study with 229 E and OC 43. Am. Rev. Respir. Dis. 1972, 105, 805–811.
- Woo, P.C.Y.; Lau, S.K.P.; Chu, C.; Chan, K.; Tsoi, H.; Huang, Y.; Wong, B.H.L.; Poon, R.W.S.; Cai, J.J.; Luk, W.; et al. Characterization and Complete Genome Sequence of a Novel Coronavirus, Coronavirus HKU1, from Patients with Pneumonia. J. Virol. 2005, 79, 884–895.
- Lau, S.K.P.; Woo, P.C.Y.; Yip, C.C.Y.; Tse, H.; Tsoi, H.W.; Cheng, V.C.C.; Lee, P.; Tang, B.S.F.; Cheung, C.H.Y.; Lee, R.A.; et al. Coronavirus HKU1 and Other Coronavirus Infections in Hong Kong. J. Clin. Microbiol. 2006, 44, 2063–2071.
- Nitsche, A.; Schweiger, B.; Ellerbrok, H.; Niedrig, M.; Pauli, G. SARS Coronavirus Detection. Emerg. Infect. Dis. 2004, 10, 1300–1303.
- Drosten, C.; Günther, S.; Preiser, W.; van der Werf, S.; Brodt, H.-R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A.M.; et al. Identification of a Novel Coronavirus in Patients with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003, 348, 1967–1976.
- Poutanen, S.M.; Low, D.E.; Henry, B.; Finkelstein, S.; Rose, D.; Green, K.; Tellier, R.; Draker, R.; Adachi, D.; Ayers, M.; et al. Identification of Severe Acute Respiratory Syndrome in Canada. N. Engl. J. Med. 2003, 348, 1995–2005.
- Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S.; Zaki, S.R.; Peret, T.; Emery, S.; Tong, S.; Urbani, C.; Comer, J.A.; Lim, W.; et al. A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003, 348, 1953–1966.
- Peiris, J.S.M.; Lai, S.T.; Poon, L.L.M.; Guan, Y.; Yam, L.Y.C.; Lim, W.; Nicholls, J.; Yee, W.K.S.; Yan, W.W.; Cheung, M.T.; et al. Coronavirus as a Possible Cause of Severe Acute Respiratory Syndrome. Lancet 2003, 361, 1319–1325.
- Killerby, M.E.; Biggs, H.M.; Midgley, C.M.; Gerber, S.I.; Watson, J.T. Middle East Respiratory Syndrome Coronavirus Transmission. Emerg. Infect. Dis. 2020, 26, 191–198.
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820.
- Al Johani, S.; Hajeer, A.H. MERS-CoV Diagnosis: An Update. J. Infect. Public Health 2016, 9, 216–219.
- Seddiq, N.; Al-Qahtani, M.; Al-Tawfiq, J.A.; Bukamal, N. First Confirmed Case of Middle East Respiratory Syndrome Coronavirus Infection in the Kingdom of Bahrain: In a Saudi Gentleman after Cardiac Bypass Surgery. Case Rep. Infect. Dis. 2017, 2017, 1–4.
- Zheng, F.; Liao, C.; Fan, Q.H.; Chen, H.B.; Zhao, X.G.; Xie, Z.G.; Li, X.L.; Chen, C.X.; Lu, X.X.; Liu, Z.S.; et al. Clinical Characteristics of Children with Coronavirus Disease 2019 in Hubei, China. Curr. Med. Sci. 2020, 40, 275–280.
- Tay, M.Z.; Poh, C.M.; Rénia, L.; MacAry, P.A.; Ng, L.F.P. The Trinity of COVID-19: Immunity, Inflammation and Intervention. Nat. Rev. Immunol. 2020, 20, 363–374.
- COVID-19 Map - Johns Hopkins Coronavirus Resource Center. Available online: (accessed on 31 May 2021).
- Toyoshima, Y.; Nemoto, K.; Matsumoto, S.; Nakamura, Y.; Kiyotani, K. SARS-CoV-2 Genomic Variations Associated with Mortality Rate of COVID-19. J. Hum. Genet. 2020, 65, 1075–1082.
- Vandenberg, O.; Martiny, D.; Rochas, O.; van Belkum, A.; Kozlakidis, Z. Considerations for Diagnostic COVID-19 Tests. Nat. Rev. Microbiol. 2020, 19, 171–183.
- Dong, Y.; Dai, T.; Wei, Y.; Zhang, L.; Zheng, M.; Zhou, F. A Systematic Review of SARS-CoV-2 Vaccine Candidates. Signal Transduct. Target. Ther. 2020, 5, 1–14.
- Elbe, S.; Buckland-Merrett, G. Data, Disease and Diplomacy: GISAID’s Innovative Contribution to Global Health. Glob. Challenges 2017, 1, 33–46.
- Agarwala, R.; Barrett, T.; Beck, J.; Benson, D.A.; Bollin, C.; Bolton, E.; Bourexis, D.; Brister, J.R.; Bryant, S.H.; Canese, K.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13.
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. NextStrain: Real-Time Tracking of Pathogen Evolution. Bioinformatics 2018, 34, 4121–4123.
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A Dynamic Nomenclature Proposal for SARS-CoV-2 Lineages to Assist Genomic Epidemiology. Nat. Microbiol. 2020, 5, 1403–1407.
- GISAID Clade and Lineage Nomenclature Aids in Genomic Epidemiology Studies of Active HCoV-19 Viruses. Available online: (accessed on 31 May 2021).
- Tang, X.; Wu, C.; Li, X.; Song, Y.; Yao, X.; Wu, X.; Duan, Y.; Zhang, H.; Wang, Y.; Qian, Z.; et al. On the Origin and Continuing Evolution of SARS-CoV-2. Natl. Sci. Rev. 2020, 7, 1012–1023.
- Hodcroft, E.B.; Hadfield, J.; Neher, R.A.; Bedford, T. Year-Letter Genetic Clade Naming for SARS-CoV-2 on Nextstain.Org. Available online: (accessed on 31 May 2021).
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733.
- Xia, S.; Lan, Q.; Su, S.; Wang, X.; Xu, W.; Liu, Z.; Zhu, Y.; Wang, Q.; Lu, L.; Jiang, S. The Role of Furin Cleavage Site in SARS-CoV-2 Spike Protein-Mediated Membrane Fusion in the Presence or Absence of Trypsin. Signal Transduct. Target. Ther. 2020, 5, 92.
- Hakim, M.S. SARS-CoV-2, Covid-19, and the Debunking of Conspiracy Theories. Rev. Med. Virol. 2021, e2222.
- Zhou, H.; Chen, X.; Hu, T.; Li, J.; Song, H.; Liu, Y.; Wang, P.; Liu, D.; Yang, J.; Holmes, E.C.; et al. A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Curr. Biol. 2020, 30, 2196–2203.e3.
- Lam, T.T.Y.; Jia, N.; Zhang, Y.W.; Shum, M.H.H.; Jiang, J.F.; Zhu, H.C.; Tong, Y.G.; Shi, Y.X.; Ni, X.B.; Liao, Y.S.; et al. Identifying SARS-CoV-2-Related Coronaviruses in Malayan Pangolins. Nature 2020, 583, 282–285.
- Zhang, T.; Wu, Q.; Zhang, Z. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr. Biol. 2020, 30, 1346–1351.e2.
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19.
- Volz, E.; Hill, V.; McCrone, J.T.; Price, A.; Jorgensen, D.; O’Toole, Á.; Southgate, J.; Johnson, R.; Jackson, B.; Nascimento, F.F.; et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell 2021, 184, 64–75.e11.
- Hou, Y.J.; Chiba, S.; Halfmann, P.; Ehre, C.; Kuroda, M.; Dinnon, K.H.; Leist, S.R.; Schäfer, A.; Nakajima, N.; Takahashi, K.; et al. SARS-CoV-2 D614G Variant Exhibits Efficient Replication Ex Vivo and Transmission in Vivo. Science 2021, 370, 1464–1468.
- PHE SARS-CoV-2 Variants of Concern and Variants under Investigation in England. Available online: (accessed on 31 May 2021).
- CDC SARS-CoV-2 Variant Classifications and Definitions. Available online: (accessed on 31 May 2021).
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated Transmissibility and Impact of SARS-CoV-2 Lineage B.1.1.7 in England. Science 2021, 372, eabg3055.
- Ferasin, L.; Fritz, M.; Ferasin, H.; Legros, V.; Leroy, E.M. Myocarditis in Naturally Infected Pets with the British Variant of COVID-19. bioRxiv 2021.
- Fujino, T.; Nomoto, H.; Kutsuna, S.; Ujiie, M.; Suzuki, T.; Sato, R.; Fujimoto, T.; Kuroda, M.; Wakita, T.; Ohmagari, N. Novel SARS-CoV-2 Variant Identified in Travelers from Brazil to Japan. Emerg. Infect. Dis. 2021, 27, 1243–1245.
- Sabino, E.C.; Buss, L.F.; Carvalho, M.P.S.; Prete, C.A.; Crispim, M.A.E.; Fraiji, N.A.; Pereira, R.H.M.; Parag, K.V.; da Silva Peixoto, P.; Kraemer, M.U.G.; et al. Resurgence of COVID-19 in Manaus, Brazil, despite High Seroprevalence. Lancet 2021, 397, 452–455.
- Naveca, F.; Souza, V.; Corado, A.; Nascimento, F.; Silva, G.; Costa, Á.; Duarte, D.; Pessoa, K.; Mejía, M.; Brandão, M.; et al. COVID-19 Epidemic in the Brazilian State of Amazonas Was Driven by Long-Term Persistence of Endemic SARS-CoV-2 Lineages and the Recent Emergence of the New Variant of Concern P.1. Res. Sq. 2021.
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D.d.S.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.S.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and Epidemiology of the P.1 SARS-CoV-2 Lineage in Manaus, Brazil. Science 2021, 372, 815–821.
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 2020, 581, 215–220.
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural Basis of Receptor Recognition by SARS-CoV-2. Nature 2020, 581, 221–224.
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310.e20.
- Tian, F.; Tong, B.; Sun, L.; Shi, S.; Zheng, B.; Wang, Z.; Dong, X.; Zheng, P. Mutation N501Y in RBD of Spike Protein Strengthens the Inter-Action between COVID-19 and Its Receptor ACE2. bioRxiv 2021.
- FDA Fact Sheet for Health Care Providers Emergency Use Authorization (EUA) of Bamlanivimab and Etesevimab. Available online: (accessed on 31 May 2021).
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135.
- FDA Fact Sheet for Health Care Providers Emergency Use Authorization (EUA) of Regen-Covtm (Casirivimab with Imdevimab). Available online: (accessed on 31 May 2021).
- Wang, P.; Casner, R.G.; Nair, M.S.; Wang, M.; Yu, J.; Cerutti, G.; Liu, L.; Kwong, P.D.; Huang, Y.; Shapiro, L.; et al. Increased Resistance of SARS-CoV-2 Variant P.1 to Antibody Neutralization. Cell Host Microbe 2021, 29, 747–751.e4.
- Edara, V.V.; Floyd, K.; Lai, L.; Gardner, M.; Hudson, W.; Piantadosi, A.; Waggoner, J.J.; Babiker, A.; Ahmed, R.; Xie, X.; et al. Infection and MRNA-1273 Vaccine Antibodies Neutralize SARS-CoV-2 UK Variant. medRxiv 2021.
- Wu, K.; Werner, A.P.; Moliva, J.I.; Koch, M.; Choi, A.; Stewart-Jones, G.B.E.; Bennett, H.; Boyoglu-Barnum, S.; Shi, W.; Graham, B.S.; et al. mRNA-1273 Vaccine Induces Neutralizing Antibodies against Spike Mutants from Global SARS-CoV-2 Variants. bioRxiv 2021.
- Collier, D.A.; De Marco, A.; Ferreira, I.A.T.M.; Meng, B.; Datir, R.; Walls, A.C.; Kemp S, S.A.; Bassi, J.; Pinto, D.; Fregni, C.S.; et al. SARS-CoV-2 B.1.1.7 Sensitivity to MRNA Vaccine-Elicited, Convalescent and Monoclonal Antibodies. medRxiv 2021.
- Altmann, D.M.; Boyton, R.J.; Beale, R. Immunity to SARS-CoV-2 Variants of Concern. Science 2021, 371, 1103–1104.
- Lule Bugembe, D.; VTPhan, M.; Ssewanyana, I.; Semanda, P.; Dhaala, B.; Nabadda, S.; Niamh, Á.O.; Rambaut, A.; Kaleebu, P.; Cotten, M. A SARS-CoV-2 Lineage A Variant (A.23.1) with Altered Spike Has Emerged and Is 1 Dominating the Current Uganda Epidemic 2. medRxiv 2021.
- Deng, X.; Garcia-knight, M.A.; Khalid, M.M.; Servellita, V.; Morris, M.K.; Sotomayor-gonzález, A.; Glasner, D.R.; Reyes, R.; Gliwa, A.S.; Reddy, N.P.; et al. Transmission, Infectivity, and Neutralization of a Spike L452R SARS-CoV-2 Variant. Cell 2021, in press.
- Yadav, P.D.; Sapkal, G.N.; Abraham, P.; Ella, R.; Deshpande, G.; Patil, D.Y.; Nyayanit, D.A.; Gupta, N.; Sahay, R.R.; Shete, A.M.; et al. Neutralization of Variant under Investigation B.1.617 with Sera of BBV152 Vaccinees. Clin. Infect. Dis. 2021, in press.
- Rahbari, R.; Moradi, N.; Abdi, M. RRT-PCR for SARS-CoV-2: Analytical Considerations. Clin. Chim. Acta 2021, 516, 1–7.
- Eftekhari, A.; Alipour, M.; Chodari, L.; Dizaj, S.M.; Ardalan, M.R.; Samiei, M.; Sharifi, S.; Vahed, S.Z.; Huseynova, I.; Khalilov, R.; et al. A Comprehensive Review of Detection Methods for SARS-CoV-2. Microorganisms 2021, 9, 232.
- Sharma, K.; Aggarwala, P.; Gandhi, D.; Mathias, A.; Singh, P.; Sharma, S.; Negi, S.S.; Bhargava, A.; Das, P.; Gaikwad, U.; et al. Comparative Analysis of Various Clinical Specimens in Detection of SARS-CoV-2 Using rRT-PCR in New and Follow up Cases of COVID-19 Infection: Quest for the Best Choice. PLoS ONE 2021, 16, e0249408.
- Tong, Y.; Bao, A.; Chen, H.; Huang, J.; Lv, Z.; Feng, L.; Cheng, Y.; Wang, Y.; Bai, L.; Rao, W.; et al. Necessity for Detection of SARS-CoV-2 RNA in Multiple Types of Specimens for the Discharge of the Patients with COVID-19. J. Transl. Med. 2020, 18, 1–8.
- Udvardi, M.K.; Czechowski, T.; Scheible, W.R. Eleven Golden Rules of Quantitative RT-PCR. Plant Cell 2008, 20, 1736–1737.
- Larionov, A.; Krause, A.; Miller, W.R. A Standard Curve Based Method for Relative Real Time PCR Data Processing. BMC Bioinform. 2005, 6, 1–16.
- Sherina, V.; McMurray, H.R.; Powers, W.; Land, H.; Love, T.M.T.; McCall, M.N. Statistical Approaches to Decreasing the Discrepancy of Non-detects in qPCR Data. bioRxiv 2017.
- Park, C.; Lee, J.; Hassan, Z.U.; Ku, K.B.; Kim, S.J.; Kim, H.G.; Park, E.C.; Park, G.S.; Park, D.; Baek, S.H.; et al. Comparison of Digital PCR and Quantitative PCR with Various SARS-CoV-2 Primer-Probe Sets. J. Microbiol. Biotechnol. 2021, 31, 358–367.
- Han, M.S.; Byun, J.H.; Cho, Y.; Rim, J.H. RT-PCR for SARS-CoV-2: Quantitative versus Qualitative. Lancet Infect. Dis. 2021, 21, 165.
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological Assessment of Hospitalized Patients with COVID-2019. Nature 2020, 581, 465–469.
- Pan, Y.; Zhang, D.; Yang, P.; Poon, L.L.M.; Wang, Q. Viral Load of SARS-CoV-2 in Clinical Samples. Lancet Infect. Dis. 2020, 20, 411–412.
- Arnaout, R.; Lee, R.A.; Lee, G.R.; Callahan, C.; Cheng, A.; Yen, C.F.; Smith, K.P.; Arora, R.; Kirby, J.E. The Limit of Detection Matters: The Case for Benchmarking Severe Acute Respiratory Syndrome Coronavirus 2 Testing. Clin. Infect. Dis. 2021, in press.
- Platts, A.E.; Johnson, G.D.; Linnemann, A.K.; Krawetz, S.A. Real-Time PCR Quantification Using a Variable Reaction Efficiency Model. Anal. Biochem. 2008, 380, 315–322.
- Lievens, A.; Van Aelst, S.; Van Den Bulcke, M.; Goetghebeur, E. Enhanced Analysis of Real-Time PCR Data by Using a Variable Efficiency Model: FPK-PCR. Nucleic Acids Res. 2012, 40, e10.
- Forootan, A.; Sjöback, R.; Björkman, J.; Sjögreen, B.; Linz, L.; Kubista, M. Methods to Determine Limit of Detection and Limit of Quantification in Quantitative Real-Time PCR (QPCR). Biomol. Detect. Quantif. 2017, 12, 1–6.
- Wikramaratna, P.S.; Paton, R.S.; Ghafari, M.; Lourenço, J. Estimating the False-Negative Test Probability of SARSCoV- 2 by RT-PCR. Eurosurveillance 2020, 25, 2000568.
- Xiao, A.T.; Tong, Y.X.; Zhang, S. False Negative of RT-PCR and Prolonged Nucleic Acid Conversion in COVID-19: Rather than Recurrence. J. Med. Virol. 2020, 92, 1755–1756.
- Erster, O.; Shkedi, O.; Benedek, G.; Zilber, E.; Varkovitzky, I.; Shirazi, R.; Shorka, D.O.; Cohen, Y.; Bar, T.; Yechieli, R.; et al. Improved Sensitivity, Safety, and Rapidity of COVID-19 Tests by Replacing Viral Storage Solution with Lysis Buffer. PLoS ONE 2021, 16, e0249149.
- Tsang, N.N.Y.; So, H.C.; Ng, K.Y.; Cowling, B.J.; Leung, G.M.; Ip, D.K.M. Diagnostic Performance of Different Sampling Approaches for SARS-CoV-2 RT-PCR Testing: A Systematic Review and Meta-Analysis. Lancet Infect. Dis. 2021, in press.
- Karligkiotis, A.; Arosio, A.; Castelnuovo, P. How to Obtain a Nasopharyngeal Swab Specimen. N. Engl. J. Med. 2020, 383, e14.
- Pinninti, S.; Trieu, C.; Pati, S.K.; Latting, M.; Cooper, J.; Seleme, M.C.; Boppana, S.; Arora, N.; Britt, W.J.; Boppana, S.B. Comparing Nasopharyngeal and Midturbinate Nasal Swab Testing for the Identification of Severe Acute Respiratory Syndrome Coronavirus 2. Clin. Infect. Dis. 2021, 72, 1253–1255.
- Mohammadi, A.; Esmaeilzadeh, E.; Li, Y.; Bosch, R.J.; Li, J.Z. SARS-CoV-2 Detection in Different Respiratory Sites: A Systematic Review and Meta-Analysis. EBioMedicine 2020, 59, 102903.
- Fani, M.; Teimoori, A.; Ghafari, S. Comparison of the COVID-2019 (SARS-CoV-2) Pathogenesis with SARS-CoV and MERS-CoV Infections. Future Virol. 2020, 15, 317–323.
- Kim, J.M.; Kim, H.M.; Lee, E.J.; Jo, H.J.; Yoon, Y.; Lee, N.J.; Son, J.; Lee, Y.J.; Kim, M.S.; Lee, Y.P.; et al. Detection and Isolation of SARS-CoV-2 in Serum, Urine, and Stool Specimens of COVID-19 Patients from the Republic of Korea. Osong Public Heal. Res. Perspect. 2020, 11, 112–117.
- Wang, W.; Xu, Y.; Gao, R.; Al, E. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844.
- Vogels, C.B.F.; Brito, A.F.; Wyllie, A.L.; Fauver, J.R.; Ott, I.M.; Kalinich, C.C.; Petrone, M.E.; Casanovas-Massana, A.; Catherine Muenker, M.; Moore, A.J.; et al. Analytical Sensitivity and Efficiency Comparisons of SARS-CoV-2 RT–QPCR Primer–Probe Sets. Nat. Microbiol. 2020, 5, 1299–1305.
- Jung, Y.; Park, G.S.; Moon, J.H.; Ku, K.; Beak, S.H.; Lee, C.S.; Kim, S.; Park, E.C.; Park, D.; Lee, J.H.; et al. Comparative Analysis of Primer-Probe Sets for RT-qPCR of COVID-19 Causative Virus (SARS-CoV-2). ACS Infect. Dis. 2020, 6, 2513–2523.
- Pillonel, T.; Scherz, V.; Jaton, K.; Greub, G.; Bertelli, C. Letter to the Editor: SARS-CoV-2 Detection by Real-Time RT-PCR. Eurosurveillance 2020, 25, 1–2.
- Klungthong, C.; Chinnawirotpisan, P.; Hussem, K.; Phonpakobsin, T.; Manasatienkij, W.; Ajariyakhajorn, C.; Rungrojcharoenkit, K.; Gibbons, R.V.; Jarman, R.G. The Impact of Primer and Probe-Template Mismatches on the Sensitivity of Pandemic Influenza A/H1N1/2009 Virus Detection by Real-Time RT-PCR. J. Clin. Virol. 2010, 48, 91–95.
- Nayar, G.; Seabolt, E.E.; Kunitomi, M.; Agarwal, A.; Beck, K.L.; Mukherjee, V.; Kaufman, J.H. Analysis and Forecasting of Global Real Time RT-PCR Primers and Probes for SARS-CoV-2. Sci. Rep. 2021, 11, 8988.
- van Kasteren, P.B.; van der Veer, B.; van den Brink, S.; Wijsman, L.; de Jonge, J.; van den Brandt, A.; Molenkamp, R.; Reusken, C.B.E.M.; Meijer, A. Comparison of Seven Commercial RT-PCR Diagnostic Kits for COVID-19. J. Clin. Virol. 2020, 128, 104412.
- Simas, A.M.; Crott, J.W.; Sedore, C.; Rohrbach, A.; Monaco, A.P.; Gabriel, S.B.; Lennon, N.; Blumenstiel, B.; Genco, C.A. Pooling for SARS-CoV2 Surveillance: Validation and Strategy for Implementation in K-12 Schools. medRxiv 2020.
- Sun, Q.; Li, J.; Ren, H.; Pastor, L.; Loginova, Y.; Madej, R.; Taylor, K.; Wong, J.K.; Zhang, Z.; Zhang, A.; et al. Saliva as a Testing Specimen with or without Pooling for SARS-CoV-2 Detection by Multiplex RT-PCR Test. PLoS ONE 2021, 16, e0243183.
- de Salazar, A.; Aguilera, A.; Trastoy, R.; Fuentes, A.; Alados, J.C.; Causse, M.; Galán, J.C.; Moreno, A.; Trigo, M.; Pérez-Ruiz, M.; et al. Sample Pooling for SARS-CoV-2 RT-PCR Screening. Clin. Microbiol. Infect. 2020, 26, 1687.e1–1687.e5.
- Barak, N.; Ben-Ami, R.; Sido, T.; Perri, A.; Shtoyer, A.; Rivkin, M.; Licht, T.; Peretz, A.; Magenheim, J.; Fogel, I.; et al. Lessons from Applied Large-Scale Pooling of 133,816 SARS-CoV-2 RT-PCR Tests. Sci. Transl. Med. 2021, 13, eabf2823.
- Cleary, B.; Hay, J.A.; Blumenstiel, B.; Harden, M.; Cipicchio, M.; Bezney, J.; Simonton, B.; Hong, D.; Senghore, M.; Sesay, A.K.; et al. Using Viral Load and Epidemic Dynamics to Optimize Pooled Testing in Resource-Constrained Settings. Sci. Transl. Med. 2021, 13, eabf1568.
- Kim, S.Y.; Lee, J.; Sung, H.; Lee, H.; Han, M.G.; Yoo, C.K.; Lee, S.W.; Hong, K.H. Pooling Upper Respiratory Specimens for Rapid Mass Screening of COVID-19 by Real-Time RT-PCR. Emerg. Infect. Dis. 2020, 26, 2469–2472.
- Wacker, M.J.; Godard, M.P. Analysis of One-Step and Two- Step Real-Time RT-PCR Using Superscript III. J. Biomol. Tech. 2005, 16, 266–271.
- Dharavath, B.; Yadav, N.; Desai, S.; Sunder, R.; Mishra, R.; Ketkar, M.; Bhanshe, P.; Gupta, A.; Redhu, A.K.; Patkar, N.; et al. A One-Step, One-Tube Real-Time RT-PCR Based Assay with an Automated Analysis for Detection of SARS-CoV-2. Heliyon 2020, 6, e04405.
- Yu, C.Y.; Chan, K.G.; Yean, C.Y.; Ang, G.Y. Nucleic Acid-Based Diagnostic Tests for the Detection SARS-CoV-2: An Update. Diagnostics 2021, 11, 53.
- Miranda, J.P.; Osorio, J.; Videla, M.; Angel, G.; Camponovo, R.; Henríquez-Henríquez, M. Analytical and Clinical Validation for RT-qPCR Detection of SARS-CoV-2 Without RNA Extraction. Front. Med. 2020, 7, 567572.
- Das Mukhopadhyay, C.; Sharma, P.; Sinha, K.; Rajarshi, K. Recent Trends in Analytical and Digital Techniques for the Detection of the SARS-Cov-2. Biophys. Chem. 2021, 270, 106538.
- Park, M.; Won, J.; Choi, B.Y.; Lee, C.J. Optimization of Primer Sets and Detection Protocols for SARS-CoV-2 of Coronavirus Disease 2019 (COVID-19) Using PCR and Real-Time PCR. Exp. Mol. Med. 2020, 52, 963–977.
- Chen, C.J.; Hsieh, L.L.; Lin, S.K.; Wang, C.F.; Huang, Y.H.; Lin, S.Y.; Lu, P.L. Optimization of the CDC Protocol of Molecular Diagnosis of COVID-19 for Timely Diagnosis. Diagnostics 2020, 10, 333.
- Bustin, S.A.; Nolan, T. RT-qPCR Testing of SARS-COV-2: A Primer. Int. J. Mol. Sci. 2020, 21, 3004.
- Wyllie, A.L.; Fournier, J.; Casanovas-Massana, A.; Campbell, M.; Tokuyama, M.; Vijayakumar, P.; Warren, J.L.; Geng, B.; Muenker, M.C.; Moore, A.J.; et al. Saliva or Nasopharyngeal Swab Specimens for Detection of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 1283–1286.
- Takeuchi, Y.; Furuchi, M.; Kamimoto, A.; Honda, K.; Matsumura, H.; Kobayashi, R. Saliva-Based PCR Tests for SARS-CoV-2 Detection. J. Oral Sci. 2020, 62, 350–351.
- Gallo, G.; La Torre, M.; Pietroletti, R.; Bianco, F.; Altomare, D.F.; Pucciarelli, S.; Gagliardi, G.; Perinotti, R. Italian Society of Colorectal Surgery Recommendations for Good Clinical Practice in Colorectal Surgery during the Novel Coronavirus Pandemic. Tech. Coloproctol. 2020, 24, 501–505.
- Mizumoto, K.; Kagaya, K.; Zarebski, A.; Chowell, G. Estimating the Asymptomatic Proportion of Coronavirus Disease 2019 (COVID-19) Cases on Board the Diamond Princess Cruise Ship, Yokohama, Japan, 2020. Eurosurveillance 2020, 25, 2000180.
- Zhu, H.; Zhang, H.; Xu, Y.; Laššáková, S.; Korabečná, M.; Neužil, P. PCR Past, Present and Future. Biotechniques 2020, 69, 317–325.
- Kojabad, A.A.; Farzanehpour, M.; Galeh, H.E.G.; Dorostkar, R.; Jafarpour, A.; Bolandian, M.; Nodooshan, M.M. Droplet Digital PCR of Viral DNA/RNA, Current Progress, Challenges, and Future Perspectives. J. Med. Virol. 2021, 93, 4182–4197.
- Sedlak, R.H.; Jerome, K.R. Viral Diagnostics in the Era of Digital Polymerase Chain Reaction. Diagn. Microbiol. Infect. Dis. 2013, 75, 1–4.
- Salipante, S.J.; Jerome, K.R. Digital PCR—An Emerging Technology with Broad Applications in Microbiology. Clin. Chem. 2020, 66, 117–123.
- Quan, P.L.; Sauzade, M.; Brouzes, E. DPCR: A Technology Review. Sensors 2018, 18, 1271.
- Kuypers, J.; Jerome, K.R. Applications of Digital PCR for Clinical Microbiology. J. Clin. Microbiol. 2017, 55, 1621–1628.
- Alteri, C.; Cento, V.; Antonello, M.; Colagrossi, L.; Merli, M.; Ughi, N.; Renica, S.; Matarazzo, E.; Ruscio, F.D.; Tartaglione, L.; et al. Detection and Quantification of SARS-CoV-2 by Droplet Digital PCR in Real-Time PCR Negative Nasopharyngeal Swabs from Suspected COVID-19 Patients. PLoS ONE 2020, 15.
- Falzone, L.; Musso, N.; Gattuso, G.; Bongiorno, D.; Palermo, C.I.; Scalia, G.; Libra, M.; Stefani, S. Sensitivity Assessment of Droplet Digital PCR for SARS-CoV-2 Detection. Int. J. Mol. Med. 2020, 46, 957–964.
- Suo, T.; Liu, X.; Feng, J.; Guo, M.; Hu, W.; Guo, D.; Ullah, H.; Yang, Y.; Zhang, Q.; Wang, X.; et al. DdPCR: A More Accurate Tool for SARS-CoV-2 Detection in Low Viral Load Specimens. Emerg. Microbes Infect. 2020, 9, 1259–1268.
- Dang, Y.; Liu, N.; Tan, C.; Feng, Y.; Yuan, X.; Fan, D.; Peng, Y.; Jin, R.; Guo, Y.; Lou, J. Comparison of Qualitative and Quantitative Analyses of COVID-19 Clinical Samples. Clin. Chim. Acta 2020, 510, 613–616.
- Liu, X.; Feng, J.; Zhang, Q.; Guo, D.; Zhang, L.; Suo, T.; Hu, W.; Guo, M.; Wang, X.; Huang, Z.; et al. Analytical Comparisons of SARS-COV-2 Detection by qRT-PCR and ddPCR with Multiple Primer/Probe Sets. Emerg. Microbes Infect. 2020, 9, 1175–1179.
- Kevadiya, B.D.; Machhi, J.; Herskovitz, J.; Oleynikov, M.D.; Blomberg, W.R.; Bajwa, N.; Soni, D.; Das, S.; Hasan, M.; Patel, M.; et al. Diagnostics for SARS-CoV-2 Infections. Nat. Mater. 2021, 20, 593–605.
- Gatti, M.; Calandri, M.; Biondo, A.; Geninatti, C.; Piatti, C.; Ruggirello, I.; Santonocito, A.; Varello, S.; Bergamasco, L.; Bironzo, P.; et al. Emergency Room Comprehensive Assessment of Demographic, Radiological, Laboratory and Clinical Data of Patients with COVID-19: Determination of Its Prognostic Value for in-Hospital Mortality. Intern. Emerg. Med. 2021, in press.
- Mostafa, H.H.; Hardick, J.; Morehead, E.; Miller, J.A.; Gaydos, C.A.; Manabe, Y.C. Comparison of the Analytical Sensitivity of Seven Commonly Used Commercial SARS-CoV-2 Automated Molecular Assays. J. Clin. Virol. 2020, 130, 104578.
- D’Cruz, R.J.; Currier, A.W.; Sampson, V.B. Laboratory Testing Methods for Novel Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2). Front. Cell Dev. Biol. 2020, 8, 1–11.
- Ganguli, A.; Mostafa, A.; Berger, J.; Aydin, M.Y.; Sun, F.; Stewart de Ramirez, S.A.; Valera, E.; Cunningham, B.T.; King, W.P.; Bashir, R. Rapid Isothermal Amplification and Portable Detection System for SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 22727–22735.
- Carter, L.J.; Garner, L.V.; Smoot, J.W.; Li, Y.; Zhou, Q.; Saveson, C.J.; Sasso, J.M.; Gregg, A.C.; Soares, D.J.; Beskid, T.R.; et al. Assay Techniques and Test Development for COVID-19 Diagnosis. ACS Cent. Sci. 2020, 6, 591–605.
- Bodulev, O.L.; Sakharov, I.Y. Isothermal Nucleic Acid Amplification Techniques and Their Use in Bioanalysis. Biochemistry 2020, 85, 147–166.
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C.W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14, 3822–3835.
- Mautner, L.; Baillie, C.K.; Herold, H.M.; Volkwein, W.; Guertler, P.; Eberle, U.; Ackermann, N.; Sing, A.; Pavlovic, M.; Goerlich, O.; et al. Rapid Point-of-Care Detection of SARS-CoV-2 Using Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP). Virol. J. 2020, 17, 1–14.
- Qian, J.; Boswell, S.A.; Chidley, C.; Lu, Z.X.; Pettit, M.E.; Gaudio, B.L.; Fajnzylber, J.M.; Ingram, R.T.; Ward, R.H.; Li, J.Z.; et al. An Enhanced Isothermal Amplification Assay for Viral Detection. Nat. Commun. 2020, 11, 1–10.
- Chaibun, T.; Puenpa, J.; Ngamdee, T.; Boonapatcharoen, N.; Athamanolap, P.; O’Mullane, A.P.; Vongpunsawad, S.; Poovorawan, Y.; Lee, S.Y.; Lertanantawong, B. Rapid Electrochemical Detection of Coronavirus SARS-CoV-2. Nat. Commun. 2021, 12, 1–10.
- Zhang, C.; Zheng, T.; Fan, H.; Zhang, T.; Han, D. Aligner-Mediated Cleavage-Based Isothermal Amplification for SARS-CoV-2 RNA Detection. ACS Appl. Bio Mater. 2021, 4, 3805–3810.
- Xing, W.; Liu, Y.; Wang, H.; Li, S.; Lin, Y.; Chen, L.; Zhao, Y.; Chao, S.; Huang, X.; Ge, S.; et al. A High-Throughput, Multi-Index Isothermal Amplification Platform for Rapid Detection of 19 Types of Common Respiratory Viruses Including SARS-CoV-2. Engineering 2020, 6, 1130–1140.
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Res. 2000, 28, E63.
- Tomita, N.; Mori, Y.; Kanda, H.; Notomi, T. Loop-Mediated Isothermal Amplification (LAMP) of Gene Sequences and Simple Visual Detection of Products. Nat. Protoc. 2008, 3, 877–882.
- Mori, Y.; Kitao, M.; Tomita, N.; Notomi, T. Real-Time Turbidimetry of LAMP Reaction for Quantifying Template DNA. J. Biochem. Biophys. Methods 2004, 59, 145–157.
- Yu, L.; Wu, S.; Hao, X.; Dong, X.; Mao, L.; Pelechano, V.; Chen, W.-H.; Yin, X. Rapid Detection of COVID-19 Coronavirus Using a Reverse Transcriptional Loop-Mediated Isothermal Amplification (RT-LAMP) Diagnostic Platform. Clin. Chem. 2020, 66, 975–977.
- Dao Thi, V.L.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.M.; Kirrmaier, D.; Freistaedter, A.; Papagiannidis, D.; Galmozzi, C.; Stanifer, M.L.; et al. A Colorimetric RT-LAMP Assay and LAMP-Sequencing for Detecting SARS-CoV-2 RNA in Clinical Samples. Sci. Transl. Med. 2020, 12, eabc7075.
- Lu, R.; Wu, X.; Wan, Z.; Li, Y.; Jin, X.; Zhang, C. A Novel Reverse Transcription Loop-Mediated Isothermal Amplification Method for Rapid Detection of Sars-Cov-2. Int. J. Mol. Sci. 2020, 21, 2826.
- Park, G.S.; Ku, K.; Baek, S.H.; Kim, S.J.; Kim, S.I.; Kim, B.T.; Maeng, J.S. Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays Targeting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Mol. Diagnostics 2020, 22, 729–735.
- Baek, Y.H.; Um, J.; Antigua, K.J.C.; Park, J.H.; Kim, Y.; Oh, S.; Kim, Y.I.; Choi, W.S.; Kim, S.G.; Jeong, J.H.; et al. Development of a Reverse Transcription-Loop-Mediated Isothermal Amplification as a Rapid Early-Detection Method for Novel SARS-CoV-2. Emerg. Microbes Infect. 2020, 9, 998–1007.
- Wei, S.; Kohl, E.; Djandji, A.; Morgan, S.; Whittier, S.; Mansukhani, M.; Hod, E.; D’Alton, M.; Suh, Y.; Williams, Z. Direct Diagnostic Testing of SARS-CoV-2 without the Need for Prior RNA Extraction. Sci. Rep. 2021, 11, 1–6.
- Lalli, M.A.; Langmade, J.S.; Chen, X.; Fronick, C.C.; Sawyer, C.S.; Burcea, L.C.; Wilkinson, M.N.; Fulton, R.S.; Heinz, M.; Buchser, W.J.; et al. Rapid and Extraction-Free Detection of SARS-CoV-2 from Saliva by Colorimetric Reverse-Transcription Loop-Mediated Isothermal Amplification. Clin. Chem. 2021, 67, 415–424.
- Pang, B.; Xu, J.; Liu, Y.; Peng, H.; Feng, W.; Cao, Y.; Wu, J.; Xiao, H.; Pabbaraju, K.; Tipples, G.; et al. Isothermal Amplification and Ambient Visualization in a Single Tube for the Detection of SARS-CoV-2 Using Loop-Mediated Amplification and CRISPR Technology. Anal. Chem. 2020, 92, 16204–16212.
- Ali, Z.; Aman, R.; Mahas, A.; Rao, G.S.; Tehseen, M.; Marsic, T.; Salunke, R.; Subudhi, A.K.; Hala, S.M.; Hamdan, S.M.; et al. ISCAN: An RT-LAMP-Coupled CRISPR-Cas12 Module for Rapid, Sensitive Detection of SARS-CoV-2. Virus Res. 2020, 288, 198129.
- Zhu, X.; Wang, X.; Han, L.; Chen, T.; Wang, L.; Li, H.; Li, S.; He, L.; Fu, X.; Chen, S.; et al. Multiplex Reverse Transcription Loop-Mediated Isothermal Amplification Combined with Nanoparticle-Based Lateral Flow Biosensor for the Diagnosis of COVID-19. Biosens. Bioelectron. 2020, 166, 112437.
- Rohaim, M.A.; Clayton, E.; Sahin, I.; Vilela, J.; Khalifa, M.E.; Al-natour, M.Q.; Bayoumi, M.; Poirier, A.C.; Branavan, M.; Tharmakulasingam, M.; et al. Artificial Intelligence-Assisted Loop Mediated Isothermal Amplification (AI-LAMP) for Rapid Detection of SARS-CoV-2. Viruses 2020, 12, 972.
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA Detection Using Recombination Proteins. PLoS Biol. 2006, 4, 1115–1121.
- Kersting, S.; Rausch, V.; Bier, F.F.; von Nickisch-Rosenegk, M. Rapid Detection of Plasmodium Falciparum with Isothermal Recombinase Polymerase Amplification and Lateral Flow Analysis. Malar. J. 2014, 13, 99.
- Lobato, I.M.; O’Sullivan, C.K. Recombinase Polymerase Amplification: Basics, Applications and Recent Advances. TrAC Trends Anal. Chem. 2018, 98, 19–35.
- Zhang, W.S.; Pan, J.; Li, F.; Zhu, M.; Xu, M.; Zhu, H.; Yu, Y.; Su, G. Reverse Transcription Recombinase Polymerase Amplification Coupled with CRISPR-Cas12a for Facile and Highly Sensitive Colorimetric SARS-CoV-2 Detection. Anal. Chem. 2021, 93, 4126–4133.
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical Validation of a Cas13-Based Assay for the Detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149.
- Arizti-Sanz, J.; Freije, C.A.; Stanton, A.C.; Petros, B.A.; Boehm, C.K.; Siddiqui, S.; Shaw, B.M.; Adams, G.; Kosoko-Thoroddsen, T.S.F.; Kemball, M.E.; et al. Streamlined Inactivation, Amplification, and Cas13-Based Detection of SARS-CoV-2. Nat. Commun. 2020, 11, 5921.
- Kim, H.S.; Abbas, N.; Shin, S. A Rapid Diagnosis of SARS-CoV-2 Using DNA Hydrogel Formation on Microfluidic Pores. Biosens. Bioelectron. 2021, 177, 113005.




