Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Dominik Łagowski | + 1641 word(s) | 1641 | 2021-06-10 10:22:14 | | | |
| 2 | Amina Yu | Meta information modification | 1641 | 2021-06-17 08:09:33 | | | | |
| 3 | Amina Yu | Meta information modification | 1641 | 2021-06-23 09:00:04 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Łagowski, D. Real-Time PCR for Detection Dermatophytes. Encyclopedia. Available online: https://encyclopedia.pub/entry/10947 (accessed on 28 February 2026).
Łagowski D. Real-Time PCR for Detection Dermatophytes. Encyclopedia. Available at: https://encyclopedia.pub/entry/10947. Accessed February 28, 2026.
Łagowski, Dominik. "Real-Time PCR for Detection Dermatophytes" Encyclopedia, https://encyclopedia.pub/entry/10947 (accessed February 28, 2026).
Łagowski, D. (2021, June 17). Real-Time PCR for Detection Dermatophytes. In Encyclopedia. https://encyclopedia.pub/entry/10947
Łagowski, Dominik. "Real-Time PCR for Detection Dermatophytes." Encyclopedia. Web. 17 June, 2021.
Copy Citation
The control of the presence of dermatophytes in herds of cattle and other species of farm animals should be routinely performed. The ongoing improvements in the field of fungal detection techniques give new scope for clinical implementations in specialized laboratories and hospitals or veterinary clinics, including the monitoring of disease and the detection of side effects of drugs and environmental risks.
dermatophytes
cattle
ringworm
Trichophyton verrucosum
Trichophyton benhamiae
1. Introduction
Dermatophytes are filamentous fungi with the ability to digest keratinized substrates, i.e., skin, hair, and nails. They are considered the major etiological agents of cutaneous superficial mycoses, often called ringworm [1][2][3][4][5]. These diseases are highly transmissible and clinically varied from mild to severe, depending on the host’s immune status, strain virulence, and other environmental factors [1][6]. The overall prevalence of ringworm in cattle is higher in countries with hotter climates and where the number of animals in the cowshed is high and the contact is closer [3][7][8][9]. Dermatophytosis is commonly considered to be a self-limiting disease, with a course of duration usually varying from 4 to 12 weeks [10]. Appropriate treatment or vaccination may shorten the period with clinical signs [11] and significantly reduce the risk of spread of infection [5].
In most countries, bovine ringworm is considered a relatively harmless disease, with little impact on production parameters and animal welfare [3][11][12]. Control measures are rarely implemented by veterinary authorities or livestock industry [10]. Nonetheless, the use of vaccine became almost routine in 1970–1980 in Hungary [13], Germany [14], Yugoslavia [15], Bulgaria [16], and Norway [17], where approximately 250 million cattle were vaccinated and the results were promising, with a drop in the percentage of infected herds from almost 100% to less than 10% in 1975 and 1% in 1984 [18]. In each of these cases, the vaccination was related to T. verrucosum dermatophytosis, although the acquired immunity was extended to trichophytoses with a different etiology [11]. Repeated administration of inactivated or attenuated preparations with a preferable 10 to 14 days interval is recommended to obtain the highest vaccine efficacy [5][11][19][20]. Interestingly, experience from natural outbreaks of ringworm in cattle has proven that the immunity is long-lasting [5][19] and that re-vaccination is not necessary [21].
An interesting issue related to ringworm in cattle is the control of the occurrence of dermatophytes in herds, and also in a carrier state. This issue is especially important in cowsheds where vaccinated and unvaccinated animals are kept. The application of conventional methods of detection and identification of dermatophytes in cattle herds has several drawbacks, as they are laborious and time-consuming and necessitate the experience of a mycological diagnostician [22][23][24]. Technologically advanced methods of dermatophyte identification developed recently include molecular methods based on PCR [25][26]. Moreover, real-time PCR techniques are increasingly frequently used in the diagnosis of human dermatomycoses, due to their high specificity and sensitivity in the detection of dermatophytes directly from clinical samples, and even if cultures assessed with conventional methods are negative [22][25][27]. These qPCR methods have so far only occasionally been used in veterinary mycology. Is it possible to use them for control studies of the presence of dermatophytes in cattle herds?
In this context, the aim of this study was to test the occurrence of dermatophytes in cattle herds using qPCR techniques and compare with the results of conventional methods.
2. Detection and Identification of Dermatophytes
The pan-dermatophyte primers in the qPCR technique (P) facilitated detection of the presence of the genetic material of dermatophytes in the sample in 100% of the cases of symptomatic animals (Table 1). Moreover, this method, when used to analyze material from the asymptomatic carrier animals, revealed that 45% of the samples contained the dermatophyte genetic material. In turn, the direct light microscopy analysis of the material (M) sampled from the clinical lesions in the cattle revealed the presence of arthrospores in all samples. In the diagnostics of the material from the asymptomatic animals, elements of the fungus were shown by the direct analysis in 34.17%. In turn, dermatophyte cultures (C) were obtained from 100% and 22.5% of the samples from the ringworm cases and asymptomatic carriage, respectively. This method allowed an identification based on the macro- and micro-morphology of the cultures (Figure 1), followed by confirmation of the results by ITS region sequencing and alignment based on the BLAST algorithm (Basic Local Alignment Search Tool), with sequences deposited in the GenBank database. The analysis of the consistency of the results obtained for the asymptomatic animals revealed that the direct examination by qPCR (P) and the microscopic evaluation (M) showed 75.93% identical results. In 50% and 22.5% of the positive results of qPCR (P) and microscopic examinations (M), respectively, culture (C) was obtained. Additionally, in 25.93% of the samples, positive results were obtained for all three methods tested.
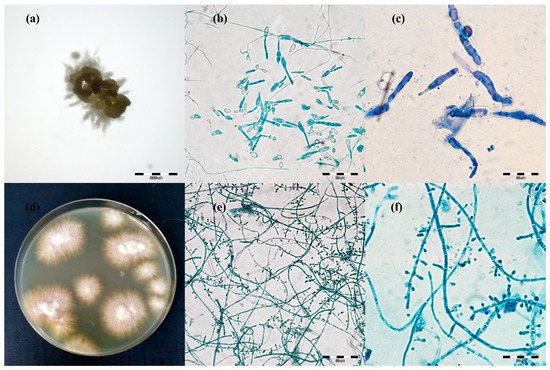
Figure 1. Micro- and macromorphology of two species of dermatophytes isolated from one outbreak of lesions from cattle. Legend: after three passages, two separate pure dermatophyte cultures were obtained: (a–c), Trichophyton verrucosum; (a), macromorphology after 21 days of incubation on Sabouraud medium at magnification 4×; (b), micromorphology after staining with lactophenol blue at magnification 400×; (c), at magnification 1000×; (d–f), Trichophyton benhamiae; (d), micromorphology (obverse); (e), micromorphology after staining with lactophenol blue at magnification 400×; (f), at magnification 1000× (pictures taken with light microscopy Olympus BX51).
Table 1. Diagnostic effectiveness of tested methods in relation to clinical material taken from symptomatic and asymptomatic dermatophyte infections.
| Type of Infection | Method (Number of Positive Animals/% of Positive Results) | Compatible (Number of Animals/% of Consistent Results) | |||||
|---|---|---|---|---|---|---|---|
| qPCR (P) | Microscopy (M) | Cultures (C) | (P)-(M) | (P)-(C) | (M)-(C) | (P)-(M)-(C) | |
| symptomatic | 40/100% | 40/100% | 40/100% | 40/100% | 40/100% | 40/100% | 40/100% |
| asymptomatic | 54/45% ST | 41/34.17% | 27/22.5% | 41/75.93% | 27/50% | 16/39.02% | 14/25.93% |
ST: significantly highest effectiveness of method (assessed by ANOVA followed by HSD Tukey test using the Statistica (Windows v13.1); StatSoft, Warsaw, Poland).
The macro- and micro-morphological image of the 21-day culture demonstrated colonies with a friable texture and a yellow or yellow-orange reverse. The size of the colonies was in the range from 3 to 7 mm. The edges of the colonies were strongly corrugated like a cauliflower surface, and the images of the obverse and the reverse were reminiscent of a star or a turbine rotor (Figure 1a). The micromorphological image on the microscope slides (Figure 21b,c) exhibited sparse, thin, and strongly elongated macroconidia. No chains of circular chlamydospores were observed. Trichophyton verrucosum was identified on the basis of morphology.
Different results were obtained for 17 samples. After 7 days of incubation of the material, the macro- and micro-morphological picture was unclear and exhibited not only macroconidia resembling the so-called rat tails, but also numerous small clavate microconidia. Such a morphological picture was not typical of T. verrucosum [3][28][29]. After the next 14 days (21st day of culture), the morphological appearance of these samples changed significantly. The size of the colonies was in the range from 8 to 12 mm. On the edge of the colony structure, there was a dense velvety light beige surface, which became suede-like, cauliflower-convex, and brown closer to the center. The micromorphological picture revealed chains of circular chlamydospores, small oval to clavate microconidia laterally inserted at the hyphae, and cigarette-shaped septated macroconidia. The morphological image appeared to be characteristic of more than one species of dermatophytes. Separate cultures were made by grafting a fragment of the mycelia from the margins and from the center of the colonies. After three passages, two separate pure dermatophyte cultures were obtained. One of them was characteristic of T. verrucosum, whereas the morphological identification of the other one revealed T. benhamiae (Figure 2d–f).
The taxonomic position of all isolates was verified based on a comparative analysis of the ITS sequences of the isolated and reference strains in the GenBank database. The ITS sequences, which were obtained for T. verrucosum isolates, in the BLAST exhibited 99% similarity to the T. verrucosum CBS365.53, and sequences of 17 isolates morphologically identified as T. benhamiae revealed 98–99% similarity to the ITS sequence of T. benhamiae CBS 809.72.
3. Enzymatic Activities
The enzymatic and haemolytic activities of the clinical isolates of dermatophytes were tested in a phenotypic assay (Table 2). In general, the enzymatic activity of the clinical isolates of dermatophytes obtained from cattle was very high. All the clinical isolates of T. verrucosum and T. benhamiae showed keratinase, phospholipase, lipase, elastase, protease, DNase, urease, and gelatinase activity (Figure 2 and Figure 3). Differences in enzymatic activity were noticeable for lipase, protease, and urease between the T. verrucosum and T. benhamiae strains. Additionally, the T. verrucosum strains produced bizonal haemolysis around the colonies, in contrast to the single and weak haemolysis caused by the T. benhamiae isolates. No differences in enzymatic activity were observed between isolates obtained from symptomatic and asymptomatic animals.
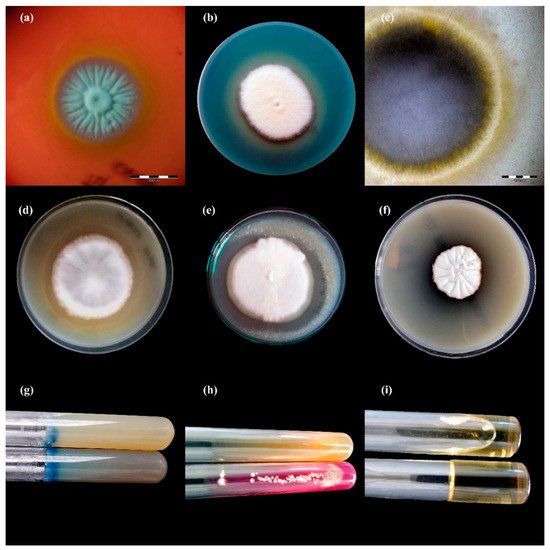
Figure 2. Enzymatic activity of Trichophyton benhamiae isolates after 21 days of incubation (picture (b,d–i): Nikon D3300, lens Nikon 18–105 mm VR; picture (a,c): Olympus SZ61, Tokio, Japan). Legend: (a), haemolysis; (b), protease; (c), lipase; (d), phospholipase; (e), elastase; (f), DNase; (g), keratinase; (h), urease; (i), gelatinase.
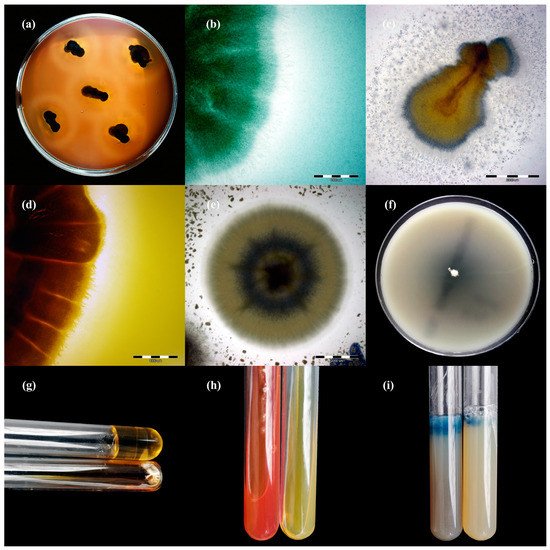
Figure 3. Enzymatic activity of Trichophyton verrucosum isolates after 21 days of incubation (picture (a,f–i): Nikon D3300, lens Nikon 18–105 mm VR; picture, (c–e): Olympus SZ61, Tokio, Japan). Legend: (a), haemolysis; (b), protease; (c), lipase; (d), phospholipase; (e), elastase; (f), DNase; (g), gelatinase; (h), urease; (i), keratinase.
Table 2. In vitro enzymatic activity of dermatophyte isolates obtained from cattle after 21 days of incubation.
| Isolates | Keratinase | Phospholipase | Lipase | Elastase | Protease | DNase | Urease | Gelatinase | Haemolysis |
|---|---|---|---|---|---|---|---|---|---|
| T. verrucosum | + | + | ++ | + | + | + | + | + | +double |
| T. benhamiae | + | + | + | + | ++ | + | ++ | + | +single |
4. Conclusions
Veterinarians agree that laboratory diagnostics and unambiguous detection of dermatophyte pathogens in cattle herds is essential for control strategies, the isolation of infected animals, and management of epidemic outbreaks. In the field of medical mycology, the dawning of the molecular diagnostics age provided the promise of rapid and reliable detection of these fungi from clinical specimens. Hence, this study aimed to evaluate the analytical specificity and clinical application of direct sample real-time PCR in comparison to the direct microscopy and culture methods. It seems that this method can be used with great success to detect and monitor cowsheds in ringworm cases and dermatophyte carriage in cattle. On the other hand, to identify the species of dermatophyte, a culture should be obtained, which can eliminate errors related to, for example, multiple-species infections.
References
- Weitzman, I.; Summerbell, R.C. The Dermatophytes. Clin. Microbiol. Rev. 1995, 8, 240–259.
- Havlickova, B.; Czaika, V.A.; Friedrich, M. Epidemiological Trends in Skin Mycoses Worldwide. Mycoses 2008, 51, 2–15.
- Gnat, S.; Łagowski, D.; Nowakiewicz, A.; Trościańczyk, A.; Zięba, P. Infection of Trichophyton Verrucosum in Cattle Breeders, Poland: A 40-Year Retrospective Study on the Genomic Variability of Strains. Mycoses 2018, 61, 681–690.
- Gnat, S.; Nowakiewicz, A.; Zięba, P. Taxonomy of Dermatophytes—The Classification Systems May Change But the Identification Problems Remain the Same. Postępy Mikrobiol. Adv. Microbiol. 2019, 58, 49–58.
- Lund, A.; Bratberg, A.M.; Næss, B.; Gudding, R. Control of Bovine Ringworm by Vaccination in Norway. Vet. Immunol. Immunopathol. 2014, 158, 37–45.
- Gnat, S.; Nowakiewicz, A.; Lagowski, D.; Czyk, A.T.; Zieba, P. Multiple-Strain Trichophyton Mentagrophytes Infection in a Silver Fox (Vulpes Vulpes) from a Breeding Farm. Med. Mycol. 2019, 57, 171–180.
- Maraki, S.; Tselentis, Y. Dermatophytoses in Crete, Greece, between 1992 and 1996. Mycoses 1998, 41, 175–178.
- Mercantini, R.; Moretto, D.; Palamara, G.; Mercantini, P.; Marsella, R. Epidemiology of Dermatophytoses Observed in Rome, Italy, between 1985 and 1993. Mycoses 1995, 38, 415–419.
- Gnat, S.; Łagowski, D.; Nowakiewicz, A.; Dyląg, M. Unusual Dermatomycoses Caused by Nannizzia Nana: The Geophilic Origin of Human Infections. Infection 2020, 48, 429–434.
- Radostits, O.; Gay, C.; Hinchcliff, K.; Constable, P. Veterinary Medicine—A Textbook of the Diseases of Cattle, Horses, Sheep, Pigs and Goats, 10th ed.; Saunders: Edinburg, TX, USA, 2006; ISBN 9780702039911.
- Wawrzkiewicz, K.; Wawrzkiewicz, J. An Inactivated Vaccine against Ringworm. Comp. Immunol. Microbiol. Infect. Dis. 1992, 15, 31–40.
- Courtellemont, L.; Chevrier, S.; Degeilh, B.; Belaz, S.; Gangneux, J.P.; Robert-Gangneux, F. Epidemiology of Trichophyton Verrucosum Infection in Rennes University Hospital, France: A 12-Year Retrospective Study. Med. Mycol. 2017, 55, 720–724.
- Horvath, Z.; Gaal, T. Results of a Trial Using LFT 130 (USSR) Live Vaccine against Ringworm Infection of Cattle. Magy. Allatorv. Lapja 1977, 32, 452–454.
- Rotermund, V.H.; Franz, H.; Hausburg, G. Erste Erfahrungen Bei Der Anwendung Der Sowjetischen Trichophytievakzine Lft-130. Mon. Vet. 1977, 32, 576–577.
- Krdzalic, P.; Stojicevic, S.; Bresjanac, D. Practical possibility for immunoprophylaxis of trichoficia in cattle in industrial breeding. Vet. Glas. 1978, 32, 343–349.
- Stankushev, K.; Duparinova, M.; Kostov, G.; Gradinarski, I. Comparative immunological studies and the determination of the epizootiological effectiveness of Soviet vaccine LTF-130 in trichophytosis. Vet. Med. Nauk. 1979, 16, 67–73.
- Aamodt, O.; Naess, B.; Sandvik, O. Vaccination of Norwegian Cattle against Ringworm. Zent. Für Veterinärmedizin Reihe B 1982, 29, 451–456.
- Sarkisov, A.K.; Nikiforov, L.T. Specific Propylaxis and Immunogenic Vaccines in the Control of Dermatophytosis. In Proceedings of the Proceddings of the 10th ISHAM Congress, Barcelona, Spain, 26 June 1988; pp. 307–312.
- Rybnikář, A.; Vrzal, V.; Chumela, J. Protective Efficacy of Vaccines against Bovine Dermatophytosis after Double and Single Vaccination. Mycoses 1998, 41, 83–86.
- Rybnikář, A.; Obořilová, E.; Hedbávný, R. Efficacy Test of Trichoben Vaccine Administered to Calves at Different Intervals between Vaccination and Re-Vaccination. Acta Vet. Brno 2008, 77, 239–243.
- Gudding, R.; Lund, A. Immunoprophylaxis of Bovine Dermatophytosis. Can. Vet. J. 1995, 36, 302–306.
- Ohst, T.; Kupsch, C.; Graser, Y.; Gräser, Y. Detection of Common Dermatophytes in Clinical Specimens Using a Simple Quantitative Real-Time TaqMan Polymerase Chain Reaction Assay. Br. J. Dermatol. 2016, 174, 602–609.
- Gnat, S.; Łagowski, D.; Nowakiewicz, A.; Dyląg, M.; Osińska, M.; Sawicki, M. Detection and Identification of Dermatophytes Based on Currently Available Methods—A Comparative Study. J. Appl. Microbiol. 2021, 130, 278–291.
- Tartor, Y.H.; Abo Hashem, M.E.; Enany, S. Towards a Rapid Identification and a Novel Proteomic Analysis for Dermatophytes from Human and Animal Dermatophytosis. Mycoses 2019, 62, 1116–1126.
- Gnat, S.; Łagowski, D.; Nowakiewicz, A.; Dyląg, M. Molecular Methods for Diagnostics of Dermatomycoses—Review of Available Techniques and Evaluation of Their Advantages and Disadvantages in Implementation for in Routine Use. Postępy Mikrobiol. Adv. Microbiol. 2019, 58, 483–494.
- Azrad, M.; Keness, Y.; Nitzan, O.; Pastukh, N.; Tkhawkho, L.; Freidus, V.; Peretz, A. Cheap and Rapid In-House Method for Direct Identification of Positive Blood Cultures by MALDI-TOF MS Technology. BMC Infect. Dis. 2019, 19, 72.
- Gnat, S.; Łagowski, D.; Nowakiewicz, A.; Osińska, M.; Kopiński, Ł. Population Differentiation, Antifungal Susceptibility, and Host Range of Trichophyton Mentagrophytes Isolates Causing Recalcitrant Infections in Humans and Animals. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 2099–2113.
- De Hoog, G.S.; Guarro, J.; Gené, J.; Figueras, M.J.; de Hoog, G.S.; Guarro, J.; Gené, J.; Figueras, M.J. Atlas of Clinical Fungi, 3rd ed.; Centraalbureau Voor Schimmelcultures (CBS): Utrecht, The Netherlands, 2019; ISBN 90-70351-43-9.
- Łagowski, D.; Gnat, S.; Nowakiewicz, A.; Osińska, M.; Trościańczyk, A.; Zięba, P. In Search of the Source of Dermatophytosis: Epidemiological Analysis of Trichophyton Verrucosum Infection in Llamas and the Breeder (Case Report). Zoonoses Public Health 2019, 66, 982–989.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.1K
Entry Collection:
Environmental Sciences
Revisions:
3 times
(View History)
Update Date:
23 Jun 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





