
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Dong-Wook Han | + 1312 word(s) | 1312 | 2021-06-02 06:14:48 | | | |
| 2 | Karina Chen | Meta information modification | 1312 | 2021-06-07 08:16:45 | | |
Video Upload Options
New virus-based sensor systems that operate on M13 bacteriophage infrastructure have attracted considerable attention. These systems can detect a range of chemicals with excellent sensitivity and selectivity. Filaments consistent with M13 bacteriophages can be ordered by highly established forms of self-assembly. This allows M13 bacteriophages to build a homogeneous distribution and infiltrate the network structure of nanostructures under mild conditions.
1. M13 Bacteriophage
Distinguishing infinitesimal amounts of chemicals or microbials in a precise and simple manner is one of the most important techniques in academia and industry. Classical sensor technology is categorized as electromagnetic (gamma radiation, optics, microwave, radio wave, and Eddy current), mechanical (sound, MEMS, and fluid), magnetic, chemical (affinity, catalytic reactions, electrochemistry, and biochemistry), nuclear (nuclear magnetic resonance), and a combination of them (opto-acoustics and membrane technology) [1][2]. Generally, the key features of sensor technology are the low cost, small size, robustness, selectivity, and sensitivity [1][3].
Recently, the use of ecofriendly materials has been one of the main points in research and industrial manufacturing. The M13 bacteriophage, which can be categorized as a natural polymer, has been a major research focus owing to its target-specific response in chemical reactions with outstanding sensitivity [4]. The bacteriophage is a human-safe viral material that only infects certain criteria of bacteria, such as Escherichia coli. [5].
The filamentous type of M13 consists of approximately 2700 copies of the spirally arranged pVIII major coat protein on the body, with 5 to 7 copies of pIII, pVI, pIX, and pVII minor coat proteins at each end. M13 has a regular form, with a length and diameter of ~880 nm and ~6.6 nm, respectively. The genome structure and protein sequences of the M13 bacteriophage are fully understood. Therefore, M13 bacteriophages are very simple to genetically engineer to achieve the desired biochemical properties [6][7][8][9]. Figure 1 shows the basic protein structures and genetic engineering pathways of M13 bacteriophages. For example, genetically modifying the pVIII protein of M13 bacteriophage to possess the tryptophan–histidine–tryptophan (WHW) peptide sequence results in it showing specific binding affinity to nitrotoluene derivatives [4][10][11][12][13][14]. Consequently, the specific binding affinity to target materials, including chemical and biological materials, by simple genetic engineering provides great possibilities to use M13 bacteriophages as the core material of sensors.
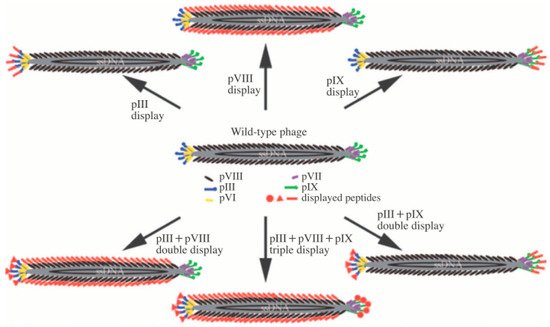
Figure 1. Basic structure of the M13 bacteriophage and the possible pathway of generic engineering. Reproduced from [9], with permission from WILEY-VCH, 2009.
M13 bacteriophages have a filamentous shape and different protein configurations at each end. One end has the pVI and pIII protein and the other has the pVII and pIX protein. By the well-defined, anisotropic structure and monodispersity caused by the natural character of M13, the material can be fabricated easily in hierarchical self-assembled structures using appropriate techniques [4][14]. The structure and behavior of M13 bacteriophages during fabrication are similar to those of liquid crystals. The bacteriophage can also be modified chemically and genetically to reveal a range of features, such as desired functionality and target-specific affiliation [11][12][13].
The target-specific reactivity character of the M13 bacteriophage makes it advantageous for sensor applications. The other strong point of the M13 bacteriophage is its ability to form higher-order dimensional structures in a specified order over self-assembly technology [13]. High-dimensional structures using natural building blocks, such as the M13 bacteriophage, were inspired by natural systems, such as the collagen bundle structure [4]. Although the imitation of a natural assembly structure on the micro scale using artificial building blocks, such as polymers or artificial nanomaterials, is progressing slowly, due to uncertainty and complexity, using natural building blocks such as M13 bacteriophage can be a solution. The filament structure and short dispersion of the M13 bacteriophage allow liquid crystal-like behavior in suspension under controlled conditions [15][16]. This liquid crystal behavior of the M13 bacteriophage can be managed mostly by the concentration of the M13 bacteriophage aqueous suspension. The crystal structure of self-assembled M13 bacteriophages shows nematic-, cholesteric-, and smectic-phase at low, medium, and high concentrations, respectively [16]. M13 is also attractive owing to its easy growth and handling characteristics. The use of natural substances as a building block makes a synthetic process possible at low cost under a non-toxic environment. The fabrication process associated with M13 bacteriophages using aqueous self-assembly technology allows easy processing without the need for organic solvents and additional processing. By using the pulling technique, concentration of M13 bacteriophage aqueous solution and polling speed of substrate are main factors in the process [17][18].
An additional outstanding benefit of M13 bacteriophages is the easy genetic modification for the specific binding affinity to definite target substances. Each coat protein can be modified genetically using phage display technology. Random phage libraries containing more than 1.0 × 1011 random peptide sequences of M13 bacteriophages are screened for specific target materials [19][20]. The binding phage on the target material is then selected and the bacteria are infected and amplified. After repeating this process to isolate the target-specific binding phage, the target-specific peptide can be recognized by DNA analysis [21][22]. Recently, the technology has developed a M13 bacteriophage that binds specifically to several inorganic resources, e.g., GaAs, GaN, Ag, Pt, Au, Pd, Ge, Ti, SiO2, quartz, CaCO3, ZnS, CdS, Co, TiO2, ZnO, CoPt, FePt, BaTiO3, CaMoO4, and hydroxyapatite [21][22][23][24][25]. The M13 bacteriophage can be made to have specific functionality by genetically engineering to carbon-based nanomaterials, such as C60, graphene, and carbon nanotubes [24][25]. Moreover, these specific binding capacities of the M13 bacteriophage can be used as templates to produce various nanostructures, such as highly monodisperse nanostructures [26][27][28]. This review introduces the current advances of M13 bacteriophage-based biosensors.
2. M13 Bacteriophage-Based Biosensor
The M13 bacteriophage has been used to improve existing sensor systems. Förster resonance energy transfer (FRET) is based on long-term bipolar interactions between excited state donors and ground state recipients. The distance of each N-terminal to the end of the peptide on the surface of a M13 bacteriophage is approximately 3.2 nm (oa) and approximately 2.3 nm (ob), respectively. Wang et al. recently reported the fabrication of FRET-based lattice probes using the M13 bacteriophage [29][30]. Owing to the regular organization of the M13 bacteriophage on the substance, a thin coating film of M13 bacteriophage can be used for surface plasmon resonance (SPR) measurements. Genetically engineered M13 bacteriophages, which possess the Arg-Gly-Asp (RGD) peptide sequence, have been used to detect the SPR signal of the cell proliferation rate and the morphology of cells [31]. The signal strength of surface-enhanced Raman spectroscopy (SERS) depends strongly on the distance between the target molecules and novel metal surface. Therefore, the controlled organization of metal nanoparticles and Raman active dye is the critical point. Cha et al. introduced an Au@Ag-core shell nanoparticle using an M13 bacteriophage as the template. In this approach, nanoparticles were functionalized by DNA-conjugated M13 bacteriophage, and showed a 75 times stronger Raman signal than DNA-functionalized nanoparticles without the M13 bacteriophage. This was mainly caused by the large number of functional moieties on pVIII protein of the M13 bacteriophage [32]. The M13 bacteriophage could also be used medically because of the strong specific reactivity to a particular reactant. Mao et al. fabricated M13–liposome–ZnPc (zinc phthalocyanine) for a more stable and upgraded cancer drug delivery system [33][34][35]. Simple genetic engineering could reveal target-specific affinity to the M13 bacteriophage. Oh et al. engineered the pVIII protein of M13 bacteriophage to have the AXXXWHWQXXDP (WHW) sequence and showed excellent binding affinity to trinitrotoluene (TNT), as shown in Figure 2a,b [36][37]. The modified M13 bacteriophage was then fabricated as a color film structure through a simple pulling technique. As shown in Figure 2c,d, the M13 bacteriophage-based color sensor could detect the gas phase of TNT down to the 300 p.p.b level and showed superior selectivity among TNT, Dinitrotoluene (DNT), and Nitrotoluene (MNT) [4].
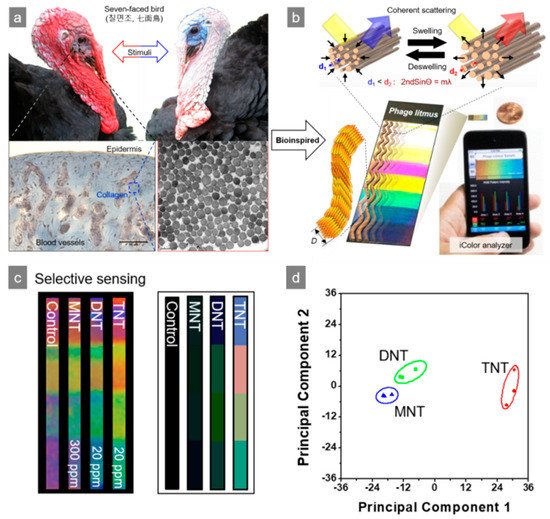
Figure 2. Nitrotoluene derivative detection using a M13 bacteriophage-based color sensor. (a) Bio-mimic structure of turkey collagen, (b) reversible color responding of a color sensor, (c) sensitivity of M13 bacteriophage-based color sensor upon exposure to nitrotoluene derivatives, and (d) principal component analysis (PCA) plot of the color changes. Reproduced from [4], with permission from Springer Nature, 2014.
References
- Andersen, P.D.; Jørgensen, B.H.; Lading, L.; Rasmussen, B. Sensor foresight—Technology and market. Technovation 2004, 24, 311–320.
- Hollingum, J. Foresight launches three sensor programmes. Sens. Rev. 1999, 19, 116–120.
- Sensors: The next wave of infotech innovation. Ten-Year Forecast Institute for the future. 1997, 115–122.
- Oh, J.-W.; Chung, W.-J.; Heo, K.; Jin, H.-E.; Lee, B.Y.; Wang, E.; Zueger, C.; Wong, W.; Meyer, J.; Kim, C.; et al. Biomimetic virus-based colorimetric sensors. Nat. Commun. 2014, 5, 3043.
- Martin-Herranz, A.; Ahmad, A.; Evans, H.M.; Ewert, K.; Schulze, U.; Safinya, C.R. Surface functionalized cationic lipid-DNA complexes for gene delivery: PEGylated lamellar complexes exhibit distinct DNA-DNA Interaction Regimes. Biophys. J. 2004, 86, 1160–1168.
- Moon, J.S.; Kim, W.G.; Kim, C.; Park, G.-T.; Heo, J.; Yoo, S.Y.; Oh, J.-W. M13 bacteriophage-based self-assembly Structures and their functional capabili-ties. Mini Rev. Org. Chem. 2015, 12, 271–281.
- Chung, W.J.; Lee, D.Y.; Yoo, S.Y. Chemical modulation of M13 bacteriophage and its functional opportunities for nanomedicine. Int. J. Nanomed. 2014, 9, 5825–5836.
- Lee, Y.J.; Yi, H.; Kim, W.-J.; Kang, K.; Yun, D.S.; Strano, M.S.; Ceder, G.; Belcher, A.M. Fabricating genetically engineered high-power lithium-ion batteries using multiple virus genes. Science 2009, 324, 1051–1055.
- Lee, Y.; Kim, J.; Yun, D.S.; Nam, Y.S.; Yang, S.-H.; Belcher, A.M. Virus-templated Au and Au-Pt core-shell nanowires and their electrocatalytic activities for fuel cell applications. Energy Sci. 2012, 5, 8328–8334.
- Mao, C.; Liu, A.; Cao, B. Virus-based chemical and biological sensing. Angew 2009, 48, 6790–6810.
- Sundar, V.C.; Yablon, A.D.; Grazul, J.L.; Ilan, M.; Aizenberg, J. Fibre-optical features of a glass sponge. Nature 2003, 424, 899–900.
- Espinosa, H.D.; Luster, A.L.; Latourte, F.J.; Loh, O.Y.; Gregoire, D.; Zavattieri, P.D. Tablet-level origin of toughening in abalone shells and translation to synthetic composite materials. Nat. Commun. 2011, 2, 173.
- Aizenberg, J.; Weaver, J.C.; Thanawala, M.S.; Sundar, V.C.; Morse, D.E.; Fratzl, P. Skeleton of euplectella sp.: Structural hierarchy from the nanoscale to the macroscale. Science 2005, 309, 275–278.
- Chung, W.-J.; Oh, J.-W.; Kwak, K.; Lee, B.Y.; Meyer, J.; Wang, E.; Hexemer, A.; Lee, S.-W. Biomimetic self-templating supramolecular structure. Naure 2011, 478, 364–368.
- Dogic, Z.; Fraden, S. Ordered phases of filamentous viruses. Curr. Opin. Colloid Interface Sci. 2006, 11, 47–55.
- Yang, S.H.; Chung, W.J.; Mcfarland, S.; Lee, S.W. Assembly of Bacteriophage into Functional Materials. Chem. Rec. 2013, 13, 43–59.
- Gimenez, S.; Lana-Villarreal, T.; Gómez, R.; Agouram, S.; Muñoz-Sanjosé, V.; Mora-Seró, I. Determination of limiting factors of photovoltaic efficiency in quantum dot sensitized solar cells: Correlation between cell performance and structural properties. J. Appl. Phys. 2010, 108, 064310.
- Karpan, V.M.; Khomyakov, P.A.; Starikov, A.A.; Giovannetti, G.; Zwierzycki, M.; Talanana, M.; Brocks, G.; van den Brink, J.; Kelly, P.J. Theoretical prediction of perfect spin filtering at interfaces between close-packed surfaces of Ni or Co and graphite or graphene. Phys. Rev. B 2008, 78, 195419.
- Smith, G.P.; Petrenko, V.A. Phage display. Chem. Rev. 1997, 97, 391–410.
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317.
- Cao, B.; Mao, C. Phage Nanobiotechnology; Petrenko, V., Smith, G.P., Eds.; RSC publishing: Cambridge, UK, 2011.
- Brissette, R.; Goldstein, N.I. Methods in Molecular Biology; Fisher, P., Ed.; Humana: Totowa, NJ, USA, 2007.
- Flynn, C.E.; Lee, S.W.; Peelle, B.R.; Belcher, A.M. Viruses as vehicles for growth, organization and assembly of materials. Acta Mater. 2003, 51, 5867–5880.
- Cui, Y.; Kim, S.N.; Jones, S.E.; Wissler, L.L.; Naik, R.R.; McAlpine, M.C. Chemical functionalization of graphene enabled by phage displayed peptides. Nano Lett. 2010, 10, 4559–4565.
- Kim, S.N.; Kuang, Z.; Slocik, J.M.; Jones, S.E.; Cui, Y.; Farmer, B.L.; McAlpine, M.C.; Naik, R.R. Preferential binding of peptides to graphene edges and planes. J. Am. Chem. Soc. 2011, 133, 14480–14483.
- Mao, C.; Solis, D.J.; Reiss, B.D.; Kottmann, S.T.; Sweeney, R.Y.; Hayhurst, A.; Georgiou, G.; Iverson, B.; Belcher, A.M. Virus-based toolkit for the directed synthesis of magnetic and semiconducting nanowires. Science 2004, 303, 213–217.
- Mio, C.; Flynn, C.E.; Hayhurst, A.; Sweeney, R.; Qi, J.; Georgiou, G.; Iverson, B.; Belcher, A.M. Viral assembly of oriented quantum dot nanowires. Proc. Natl. Acad. Sci. USA 2003, 100, 6946–6951.
- Huang, Y.; Chiang, C.-Y.; Lee, S.K.; Gao, Y.; Hu. E., L.; De Yoreo, J.; Belcher, A.M. Programmable assembly of nanoarchitectures using genetically engineered viruses. Nano Lett. 2005, 5, 1429–1434.
- Deo, S.; Godwin, H.A. A selective, ratiometric fluorescent sensor for Pb2+. J. Am. Chem. Soc. 2000, 122, 174–175.
- Chen, L.; Wu, Y.; Lin, Y.; Wang, Q. Virus-templated FRET platform for the rational design of ratiometric fluorescent nanosensors. Chem. Commun. 2015, 51, 10190–10193.
- Yoo, S.Y.; Oh, J.-W.; Lee, S.W. Phage-chips for novel optically readable tissue engineering assays. Langmuir 2012, 28, 2166–2172.
- Lee, J.H.; Xu., P.F.; Domaille, D.W.; Choi, C.; Jin, S.; Cha, J.N. M13 Bacteriophage as materials for amplified surface enhanced Raman scattering protein sensing. Adv. Funct. Mater. 2014, 24, 2079–2084.
- Kawakami, K.; Nishihara, Y.; Hirano, K. Effect of hydrophilic polymers on physical stability of liposome dispersions. J. Phys. Chem. B 2001, 105, 2374–2385.
- Ruysschaert, T.; Germain, M.; Gomes, J.F.; Fournier, D.; Sukhorukov, G.B.; Meier, W.; Winterhalter, M. Liposome-based nanocapsules. IEEE Trans. Nanobiosci. 2004, 3, 49–55.
- Ngweniform, P.; Abbineni, G.; Cao, B.; Mao, C. Self-assembly of drug-loaded liposomes on genetically engineered target-recognizing M13 phage: A novel nanocarrier for targeted drug delivery. Small 2009, 5, 1963–1969.
- Jaworski, J.W.; Raorane, D.; Huh, J.H.; Majumdar, A.; Lee, S.-W. Evolutionary screening of biomimetic coatings for selective detection of explosives. Langmuir 2008, 24, 4938–4943.
- Jin, H.; Won, N.; Ahn, B.; Kwang, J.; Heo, K.; Oh, J.-W.; Sun, Y.; Cho, S.G.; Lee, S.-W.; Kim., S. Quantum dot-engineered M13 virus layer-by-layer composite films for highly selective and sensitive turn-on TNT sensors. Chem. Commun. 2013, 49, 6045–6047.




