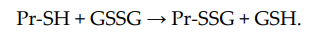| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Elena Kalinina | + 2550 word(s) | 2550 | 2021-01-22 07:33:20 | | | |
| 2 | Lily Guo | -1 word(s) | 2549 | 2021-02-01 08:55:51 | | |
Video Upload Options
Glutathione refers to the small molecule protein consisting of three amino acids - glycine, cysteine, and glutamine.
1. Introduction
Redox-dependent processes largely determine cell viability, participating in the regulation of division, bioenergetics, and programmed death. The cellular redox status is characterized by low-molecular-weight indicators (GSH, NADH). The change in their oxidized/reduced form ratio occurs as a reaction to changes in reactive oxygen and nitrogen species (RONS) levels and, so, they can play the role of a trigger in the redox-dependent regulation of cellular processes. Undoubtedly, such an important trigger role is played by glutathione (γ-glutamyl-l-cysteinylglycine, GSH), a water-soluble tripeptide consisting of the amino acids l-glutamate, l-cysteine, and glycine, which is widely present in both eukaryotes and prokaryotes[1][2][3]. GSH is less susceptible to oxidation than Cys, which makes it the most suitable for maintaining intracellular redox status[1]. The presence of a γ-peptide bond at the Glu residue protects GSH from the action of peptidases, while the SH group at the Cys residue makes GSH a good electron donor, allowing it to participate in reactions with strong electrophiles.
Normally, the ratio of reduced glutathione (GSH) to oxidized glutathione (GSSG), GSH/GSSG—which characterizes the cellular redox status—is 100/1 in the cytoplasm, 10/1 in mitochondria, and 3/1 to 1 in the endoplasmic reticulum[4]. This ratio varies depending on the physiological state of cells, such as proliferation, differentiation, or apoptosis, and the consequences of its disturbance are significant changes in cellular signaling reactions. This role of GSH/GSSG is largely due to its regulatory effect on the functional activities of protein thiols[5][6][7].
Although Cys residues in mammalian proteins do not exceed 3%[6], they are highly sensitive to oxidative modification, which significantly affects the functioning of proteins, as thiol groups play a significant role in the formation of protein tertiary and quaternary structures and enzyme active sites. The pKa value of most SH groups of cellular proteins is more than 8.0, which keeps thiol groups predominantly protonated at physiological pH values (pH 7.0–7.4)[8][9]. However, in proteins, in the immediate vicinity of the basic amino acid residues (histidine, lysine, and arginine), the pKa of the SH groups decreases (usually to 5.0–7.0) and these thiols dissociate at physiological pH. The resulting thiolate anions (Pr-S−) are effective nucleophiles and have high activity with respect to electrophilic targets[10][11][12]. The reactivity of SH groups and the functional activity of proteins are largely regulated by S-glutathionylation and S-nitrosylation[6][13][14][15][16].
Under S-glutathionylation, GSH can bind to the cysteinyl residues of proteins through the creation of reversible disulfide bonds, depending on the cysteine position and redox potential [12][13]. This post-translational modification to the protein can lead to enhanced or suppressed activity, may prevent protein degradation by proteolysis or sulfhydryl overoxidation, and plays an important role in cellular signaling. At present, the dual role of S-glutathionylation in maintaining cellular homeostasis and participating in various pathological processes may be indicated[14][17].
Under S-nitrosylation, NO is covalently attached to the SH group of a cysteine residue and, as a consequence, can cause alterations in the cellular function of a variety of proteins[18][19]. S-nitrosoglutathione, formed as the result of GSH S-nitrosylation, serves as a NO reservoir and can transfer NO groups to new cysteine residues in transnitrosylation reactions[20][21].
The GSH/GSSG ratio can be considered a key redox sensor, which determines the redox-dependent alteration of the protein functional activity through such significant post-translational modifications as S-glutathionylation and S-nitrosylation. The modulation of the activity of these reactions in response to a change in the GSH/GSSG ratio, as a result of an increase or decrease in RONS levels, provides a significant contribution to cell functional adaptation to redox changes of the environment [22][23][24][25].
In this review, we analyze data concerning the roles of GSH and GSH/GSSG in the redox modulation of S-glutathionylation and S-nitrosylation and their relationship for the maintenance of cell viability.
2. Glutathione and Protein S-Glutathionylation
A widespread form of cysteine modification is S-glutathionylation—the reversible formation of protein mixed disulfides with GSH (Pr-SSG)—which occurs in the cell under physiological conditions and oxidative stress, both spontaneously and enzymatically. The S-glutathionylation of proteins suggests the possible involvement of this post-translational modification in cellular signaling and the redox regulation of protein functions[6][13][24]. In addition to the potential regulatory role, S-glutathionylation can serve as a means of GSH storage, as well as protection from the irreversible oxidation of protein thiol groups under stress conditions[26], often due to a temporary loss of primary protein activity as, if the modified sulfhydryl group of a protein is functionally critical, S-glutathionylation can render the protein inactive or alter its activity, ultimately disrupting cellular functions[27]. In addition, this reaction can affect a change in conformation and/or charge, which can modify the function of the protein, as the attachment of GSH introduces an additional negative charge at the expense of the glutamic acid residue.
Non-enzymatic S-glutathionylation reactions can occur during thiol-disulfide exchange, through the participation of protein thiol (Pr-SH) and oxidized glutathione GSSG:
The equilibrium constant of the reaction Kmix is expressed by the ratio [Pr-SSG]·[GSH]/[Pr-SH]·[GSSG], where the extent of S-glutathionylated proteins ([Pr-SSG]/[Pr-SH]) strongly depends on the local ratio [GSH]/[GSSG][28][29]. The S-glutathionylation of most proteins with typical redox potential (Kmix~1) by about 50% can occur when this ratio drops very dramatically (i.e., from 100:1 to 1:1). These extreme conditions are rare in vivo. Therefore, for most proteins, the spontaneous formation of Pr-SSG—as a result of the exchange of Pr-SH and GSSG—is not common and, as a rule, takes place under pathological conditions[30]. In addition, a variant of thiol-disulfide exchange between Pr-SH and a protein which is already S-glutathionylated (Pr′-SSG) is possible:
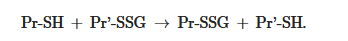
S-glutathionylation can occur when Pr-SH or GSH reacts with an oxidized derivative of the protein cysteine residue; for example, sulfenic acid (-SOH), thiyl radical (-S•), or S-nitrosyl (-SNO) group. Thus, when Pr-SH is oxidized with, for example, H2O2, sulfenic acid (Pr-SOH) is formed and then quickly reacts with GSH to form Pr-SSG:
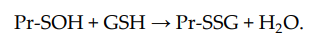
Sulfenic acid is unstable and can undergo further oxidation to sulfinic acid (Pr-SO2H) and eventually to sulfonic acid (Pr-SO3H), the formation of which, as a rule, leads to the irreversible deactivation of the protein. Thus, the S-glutathionylation of sulfenic acid can prevent the oxidation of protein cysteine residues[6][25]. Under physiological conditions, the intracellular level of H2O2 is in the sub-micromolar range (10−9–10−7 M) [31]. Therefore, in vivo spontaneous S-glutathionylation proceeds rather slowly by this mechanism.
The formation of S-glutathionylated protein is also possible, due to its interaction with GSSG in the form of sulfenic acid (GSOH):
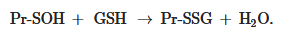
Thus, it is obvious that the S-glutathionylation of proteins can occur spontaneously; however, the rate and extent of this process increases with the participation of enzymes, among which glutathione transferase isoform P1-1 (GSTP1-1) plays the greatest role[32][33]. GSTP1-1 has been shown to facilitate S-glutathionylation for a number of proteins, including peroxiredoxin 6 (Prx6)[34][35], aldose reductase[36], actin[37], histone H3[38], 5’AMP-activated protein kinase (AMPK)[32], estrogen receptor α[39], heat shock protein BiP, protein disulfide isomerase (PDI), calnexin, calreticulin, and sarcoplasmic reticulum Ca2+-ATPase (SERCA)[33].
During the reaction, GSTP1-1 binds GSH in the active center and decreases the pKa of the GSH cysteine residue from 9.2 to 6.3[40], deprotonates it with the participation of Tyr7, forming a thiolate anion (GS−), which is transferred to the cysteine residue in the substrate. In cells with the Tyr7 GSTP1-1 mutation, a decrease in the total content of S-glutathionylated proteins has been observed upon treatment with GSSG, the mimetic of oxidized glutathione NOV-002 and diazeniumdiolate-based NO-donor prodrug PABA/NO[33]. An example is the S-glutathionylation of Prx6 from the 1-Cys groups of peroxiredoxins. As a result of human Prx6 peroxidase activity, the Cys47 in the active center is oxidized to sulfenic acid; this deprives it of activity; as for the reduction of which, a second thiol is required to form a mixed disulfide, then a sulfhydryl group. However, the availability of the sulfenic group is low, due to the peculiarities of the globular structure of Prx6. Prx6 activation occurs during the formation of a heterodimer with GSTP1-1, which promotes the S-glutathionylation of Cys47 Prx6. The conformational changes of the heterodimer occur, providing the formation of a disulfide bond between Cys47 GSTP1-1 and Cys47 Prx6, followed by the reduction of disulfide with the participation of GSH and the regeneration of Cys47 Prx6[34].
The enzymatic S-glutathionylation of AMP-activated protein kinase (AMPK) occurs not only with the participation of GSTP1-1, but also GSTM1-1 in the absence of strong oxidants (i.e., under conditions which are similar to physiological oxidation). S-glutathionylation occurs at the Cys299 and Cys304 residues and causes conformational changes that activate the kinase activity of human AMPK[32].
The ability of S-glutathionylation was found in the enzyme glyoxylase 2 (Glo2). Glo2 hydrolyzes S-d-lactoylglutathione to glutathione and lactic acid, while GS− is formed in the active center of Glo2, similar to GSTP1-1[41]. It has been established that actin and malate dehydrogenase can serve as substrates for S-glutathionylation by Glo2[42].
S-glutathionylation is a reversible post-translational modification and, as a rule, deglutathionylation proceeds with the participation of enzymes and is more carefully regulated, in comparison with S-glutathionylation. Glutaredoxin (Grx) is one of the most effective and well-studied enzymes, which reduces Pr-SSG. In the traditional classification, they are divided into mono- and dithiol Grx, depending on whether one or two cysteine residues, respectively, are in the active center. The role of dithiol Grxs is mainly considered in the regulation of reversible S-glutathionylation [26][43][44].
Mammalian dithiol Grxs, Grx1 and Grx2, are found in many cellular compartments; however, Grx1 is mainly present in the cytosol (~1 μM) and mitochondrial intermembrane space (~0.1 μM), while Grx2 is localized mainly in the mitochondrial matrix (~1 μM) [45][46]. Being thiol oxidoreductases, Grx1 and Grx2 contain the CXXC motif (CysN-XX-CysC; CPYC in Grx1 and CSYC in Grx2) in the active site. In addition, they are characterized by the presence of a thioredoxin fold, consisting of four β-sheets surrounded by three α-helices, and a site responsible for stabilizing GSH. Grx uses GSH as a co-substrate for the reduction of Pr-SSG mixed disulfides.
It should be noted that, depending on the value of the GSH/GSSG ratio, Grx can not only carry out deglutathionylation but, on the contrary, may promote S-glutathionylation (Figure 1). Grx2 functions as a glutathionylation enzyme under a decrease of GSH/GSSG and an increase in the level of Н2О2 (e.g., in relation to the respiratory complex I) whereas, at a high level of GSH/GSSG, and low concentrations of Н2О2, Grx2 has a deglutathionylating activity [47]. The putative mechanism of S-glutathionylation proceeds in several stages: first, there is a nucleophilic attack of the disulfide bond GSSG by the thiolate anion Grx-S−, along with the formation of the glutathionylated intermediate Grx-SSG, from which the activated cationic radical [GS]+ is transferred to the target protein with the formation of Pr-SSG, while Grx is again capable of catalyzing the reaction. For this process, the possibility of the reversible formation of Grx-S2 from Grx-SSG is also noted[48].
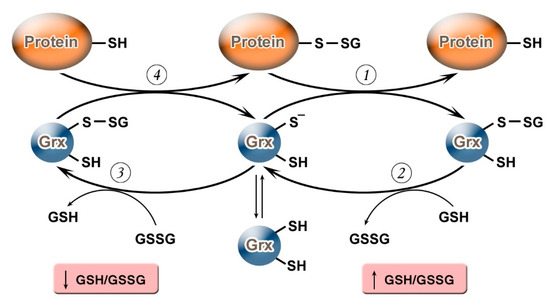
Figure 1. Glutaredoxin catalytic mechanism in dependence of GSH/GSSH ratio. Under an increase in GSH/GSSG, Grx can catalyze the deglutathionylation of proteins. The glutathionylated sulfur moiety of the protein–SSG is attacked by the thiolate anion of the enzyme (Grx-S−), forming the covalent enzyme intermediate (GRx–SSG) and releasing the reduced protein–SH as the first product (1). The second rate-determining step involves the reduction of Grx–SSG by GSH to produce glutathione disulfide (GSSG) as the second product, recycling the reduced enzyme (Grx–S−) (2). Under conditions of decreased GSH/GSSG ratio, Grx can catalyze S-glutathionylation of proteins. The S-glutathionylated Grx (Grx–SSG), formed in reaction with GSSG (3), reacts with a protein to create S-glutathionylated protein (protein–SSG) (4).
In addition to Grx, the ability to catalyze deglutathionylation has been observed in some other enzymes (Table 1).
The isoform of glutathione transferase, GSTO1-1, has the ability to catalyze protein deglutathionylation[54][55]. The isozyme is structurally similar to Grx, including Trx-like folding and a glutathione binding site, where it can form a disulfide bond with a conserved cysteine residue in the active site[53]. Other GST isoforms, including GSTA, GSTM, GSTP, GSTT, GSTS, and GSTZ, in contrast, have catalytic tyrosine or serine residues. In addition, GSTO1-1 has a relatively accessible pocket in the active site, which can potentially accommodate a protein or peptide as a substrate[54][64]. GSTO1-1 catalyzes Grx-like protein deglutathionylation in two similar stages: in the first, the Cys32 of the active site in human GSTO1-1 interacts with Pr-SSG, resulting in reduced Pr-SH and mixed disulfide GSTO1-1-Cys32S-SG, which is deglutathionylated with the participation of GSH to form GSSG and functional active GSTO1-1, which is capable of catalyzing the deglutathionylation of the next protein substrate[54]. The question of the role that GSTO1-1 plays in the S-glutathionylation of proteins remains open [53].
The process of deglutionylation is also carried out with the participation of the main members of the Trx family, which play essential roles in maintaining cellular redox homeostasis. Thioredoxins (Trx) 1 and 2 restore disulfide bonds in proteins. This process involves two cysteine residues of the Trx active site (Cys-X-X-Cys), where the disulfide bond is transferred from the substrate protein to Trx. Then, the oxidized Trx is reduced by the NADPH-dependent Trx reductase (TrxR)[65]. In addition, using the mechanism of dithiol reduction, Trxs are able to carry out deglutathionylation without the participation of GSH, which has been shown for glyceraldehyde-3-phosphate dehydrogenase, Prx3, 20S proteasome, and NOS3[59][60][61][62]; however, the exact mechanism of deglutathionylation has not yet been determined.
The ability to deglutathionate proteins has also been observed in protein disulfide isomerases (PDIs), which are also included in the Trx family[63]. However, the significance of the PDI contribution to this process is not yet clear, as their main function is the exchange of the disulfide bonds of PDIs and target proteins. PDIs are enzymes of the endoplasmic reticulum, which are specifically responsible for protein folding through the oxidation of newly formed proteins and isomerization of proteins with improperly formed disulfide bonds, achieving the formation of their native structure. Moreover, PDIs can be secreted by the cell or associated with the cell surface to maintain proteins in a reduced state[62].
Sulfiredoxin (Srx), for which the ability to reduce the cysteine residue oxidized to sulfinic acid in the active site of typical 2-Cys perxiredoxins (2-Cys Prx) was originally established, is also capable of deglutathionylation, in relation to at least the Prx isoforms[13][56][57], actin, and tyrosine protein phosphatase 1B[58]. For example, it has been shown in vitro that human Prx1 can be S-glutathionylated at three out of four cysteine residues—Cys52, Cys173, and Cys83—and deglutathionylation at Cys83 and Cys173 is catalyzed by Srx, while deglutathionylation at Cys52 is carried out by Grx[56]. The mechanism of Srx-catalyzed deglutathionylation has not yet been fully determined. The data indicate that it proceeds by a mechanism similar to that catalyzed by Grx through the formation of the Srx-SSG intermediate glutathionylated at the conservative Cys99 residue[56]. Srx-catalyzed deglutathionylation appears to have broad substrate specificity. In HEK293 cells transfected with Srx, a decrease in the total content of S-glutathionylated proteins formed under conditions of nitrosative stress after treatment with the nitric oxide donor PABA/NO has been demonstrated[58].
References
- Dwivedi, D.; Megha, K.; Mishra, R.; Mandal, P.K. Glutathione in Brain: Overview of Its Conformations, Functions, Bio-chemical Characteristics, Quantitation and Potential Therapeutic Role in Brain Disorders. Neurochem. Res. 2020, 45, 1461–1480, doi:10.1007/s11064-020-03030-1.
- Lv, H.; Zhen, C.; Liu, J.; Yang, P.; Hu, L.; Shang, P. Unraveling the Potential Role of Glutathione in Multiple Forms of Cell Death in Cancer Therapy. Oxid. Med. Cell. Longev. 2019, 2019, 3150145, doi:10.1155/2019/3150145.
- Paul, B.D.; Sbodio, J.I.; Snyder, S.H. Cysteine Metabolism in Neuronal Redox Homeostasis. Trends Pharmacol. Sci. 2018, 39, 513–524, doi:10.1016/j.tips.2018.02.007.
- Moriarty-Craige, S.E.; Jones, D.P. Extracellular thiols and thiol/disulfide redox in metabolism. Annu. Rev. Nutr. 2004, 24, 481–509, doi:10.1146/annurev.nutr.24.012003.132208.
- Giustarini, D.; Colombo, G.; Garavaglia, M.L.; Astori, E.; Portinaro, N.M.; Reggiani, F.; Badalamenti, S.; Aloisi, A.M.; San-tucci, A.; Rossi, R.; et al. Assessment of glutathione/glutathione disulphide ratio and S-glutathionylated proteins in human blood, solid tissues, and cultured cells. Free Radic. Biol. Med. 2017, 112, 360–375, doi:10.1016/j.freeradbiomed.2017.08.008.
- Musaogullari, A.; Chai, Y.C. Redox Regulation by Protein S-Glutathionylation: From Molecular Mechanisms to Implica-tions in Health and Disease. Int. J. Mol. Sci. 2020, 21, 8113, doi:10.3390/ijms21218113.
- Bjorklund, G.; Tinkov, A.A.; Hosnedlova, B.; Kizek, R.; Ajsuvakova, O.P.; Chirumbolo, S.; Skalnaya, M.G.; Peana, M.; Dadar, M.; El-Ansary, A.; et al. The role of glutathione redox imbalance in autism spectrum disorder: A review. Free Radic. Biol. Med. 2020, 160, 149–162, doi:10.1016/j.freeradbiomed.2020.07.017.
- Giles, G.I.; Tasker, K.M.; Jacob, C. Hypothesis: The role of reactive sulfur species in oxidative stress. Free Radic. Biol. Med. 2001, 31, 1279–1283, doi:10.1016/s0891-5849(01)00710-9.
- Shi, Y.; Carroll, K.S. Activity-Based Sensing for Site-Specific Proteomic Analysis of Cysteine Oxidation. Acc. Chem. Res. 2020, 53, 20–31, doi:10.1021/acs.accounts.9b00562.
- Grek, C.L.; Zhang, J.; Manevich, Y.; Townsend, D.M.; Tew, K.D. Causes and consequences of cysteine S-glutathionylation. J. Biol. Chem. 2013, 288, 26497–26504, doi:10.1074/jbc.R113.461368.
- Bechtel, T.J.; Weerapana, E. From structure to redox: The diverse functional roles of disulfides and implications in disease. Proteomics 2017, 17, 1600391, doi:10.1002/pmic.201600391.
- Benhar, M. Oxidants, Antioxidants and Thiol Redox Switches in the Control of Regulated Cell Death Pathways. Antioxid. (Basel) 2020, 9, 309, doi:10.3390/antiox9040309.
- Zhang, J.; Ye, Z.W.; Singh, S.; Townsend, D.M.; Tew, K.D. An evolving understanding of the S-glutathionylation cycle in pathways of redox regulation. Free Radic. Biol. Med. 2018, 120, 204–216, doi:10.1016/j.freeradbiomed.2018.03.038.
- Mailloux, R.J. Protein S-glutathionylation reactions as a global inhibitor of cell metabolism for the desensitization of hy-drogen peroxide signals. Redox Biol. 2020, 32, 101472, doi:10.1016/j.redox.2020.101472.
- Shi, X.; Qiu, H. Post-Translational S-Nitrosylation of Proteins in Regulating Cardiac Oxidative Stress. Antioxid. (Basel) 2020, 9, 1051, doi:10.3390/antiox9111051.
- Zhang, Y.; Deng, Y.; Yang, X.; Xue, H.; Lang, Y. The Relationship Between Protein S-Nitrosylation and Human Diseases: A Review. Neurochem. Res. 2020, 45, 2815–2827, doi:10.1007/s11064-020-03136-6.
- Lermant, A.; Murdoch, C.E. Cysteine Glutathionylation Acts as a Redox Switch in Endothelial Cells. Antioxid. (Basel) 2019, 8, 315, doi:10.3390/antiox8080315.
- Mishra, D.; Patel, V.; Banerjee, D. Nitric Oxide and S-Nitrosylation in Cancers: Emphasis on Breast Cancer. Breast Cancer (Auckl) 2020, 14, 1178223419882688, doi:10.1177/1178223419882688.
- Begara-Morales, J.C.; Sanchez-Calvo, B.; Chaki, M.; Valderrama, R.; Mata-Perez, C.; Padilla, M.N.; Corpas, F.J.; Barroso, J.B. Antioxidant Systems are Regulated by Nitric Oxide-Mediated Post-translational Modifications (NO-PTMs). Front. Plant Sci. 2016, 7, 152, doi:10.3389/fpls.2016.00152.
- Choi, M.S. Pathophysiological Role of S-Nitrosylation and Transnitrosylation Depending on S-Nitrosoglutathione Levels Regulated by S-Nitrosoglutathione Reductase. Biomol. Ther. (Seoul) 2018, 26, 533–538, doi:10.4062/biomolther.2018.179.
- Arora, D.; Jain, P.; Singh, N.; Kaur, H.; Bhatla, S.C. Mechanisms of nitric oxide crosstalk with reactive oxygen species scav-enging enzymes during abiotic stress tolerance in plants. Free Radic. Res. 2016, 50, 291–303, doi:10.3109/10715762.2015.1118473.
- Belcastro, E.; Gaucher, C.; Corti, A.; Leroy, P.; Lartaud, I.; Pompella, A. Regulation of protein function by S-nitrosation and S-glutathionylation: Processes and targets in cardiovascular pathophysiology. Biol. Chem. 2017, 398, 1267–1293, doi:10.1515/hsz-2017-0150.
- Weise-Cross, L.; Resta, T.C.; Jernigan, N.L. Redox Regulation of Ion Channels and Receptors in Pulmonary Hypertension. Antioxid. Redox Signal. 2019, 31, 898–915, doi:10.1089/ars.2018.7699.
- Dominko, K.; Dikic, D. Glutathionylation: A regulatory role of glutathione in physiological processes. Arh. Hig. Rada Toksikol. 2018, 69, 1–24, doi:10.2478/aiht-2018-69-2966.
- Moldogazieva, N.T.; Mokhosoev, I.M.; Feldman, N.B.; Lutsenko, S.V. ROS and RNS signalling: Adaptive redox switches through oxidative/nitrosative protein modifications. Free Radic. Res. 2018, 52, 507–543, doi:10.1080/10715762.2018.1457217.
- Young, A.; Gill, R.; Mailloux, R.J. Protein S-glutathionylation: The linchpin for the transmission of regulatory information on redox buffering capacity in mitochondria. Chem. Biol. Interact. 2019, 299, 151–162, doi:10.1016/j.cbi.2018.12.003.
- Nulton–Persson, A.C.; Starke, D.W.; Mieyal, J.J.; Szweda, L.I. Reversible inactivation of alpha-ketoglutarate dehydrogenase in response to alterations in the mitochondrial glutathione status. Biochemistry 2003, 42, 4235–4242, doi:10.1021/bi027370f.
- Gilbert, H.F. Thiol/disulfide exchange equilibria and disulfide bond stability. Methods Enzymol. 1995, 251, 8–28, doi:10.1016/0076-6879(95)51107-5.
- Gilbert, H.F. Molecular and cellular aspects of thiol-disulfide exchange. Adv. Enzymol. Relat. Areas Mol. Biol. 1990, 63, 69–172, doi:10.1002/9780470123096.ch2.
- Rashdan, N.A.; Shrestha, B.; Pattillo, C.B. S-glutathionylation, friend or foe in cardiovascular health and disease. Redox Biol. 2020, 37, 101693, doi:10.1016/j.redox.2020.101693.
- Arbault, S.; Pantano, P.; Sojic, N.; Amatore, C.; Best-Belpomme, M.; Sarasin, A.; Vuillaume, M. Activation of the NADPH oxidase in human fibroblasts by mechanical intrusion of a single cell with an ultramicroelectrode. Carcinogenesis 1997, 18, 569–574, doi:10.1093/carcin/18.3.569.
- Klaus, A.; Zorman, S.; Berthier, A.; Polge, C.; Ramirez, S.; Michelland, S.; Seve, M.; Vertommen, D.; Rider, M.; Lentze, N.; et al. Glutathione S-transferases interact with AMP-activated protein kinase: Evidence for S-glutathionylation and activation in vitro. PLoS ONE 2013, 8, e62497, doi:10.1371/journal.pone.0062497.
- Ye, Z.W.; Zhang, J.; Ancrum, T.; Manevich, Y.; Townsend, D.M.; Tew, K.D. Glutathione S-Transferase PMediated Protein S-Glutathionylation of Resident Endoplasmic Reticulum Proteins Influences Sensitivity to Drug-Induced Unfolded Protein Response. Antioxid. Redox Signal. 2017, 26, 247–261, doi:10.1089/ars.2015.6486.
- Ralat, L.A.; Manevich, Y.; Fisher, A.B.; Colman, R.F. Direct evidence for the formation of a complex between 1-cysteine peroxiredoxin and glutathione S-transferase pi with activity changes in both enzymes. Biochemistry 2006, 45, 360–372, doi:10.1021/bi0520737.
- Zhou, S.; Sorokina, E.M.; Harper, S.; Li, H.; Ralat, L.; Dodia, C.; Speicher, D.W.; Feinstein, S.I.; Fisher, A.B. Peroxiredoxin 6 homodimerization and heterodimerization with glutathione S-transferase pi are required for its peroxidase but not phos-pholipase A2 activity. Free Radic. Biol. Med. 2016, 94, 145–156, doi:10.1016/j.freeradbiomed.2016.02.012.
- Townsend, D.M.; Manevich, Y.; He, L.; Hutchens, S.; Pazoles, C.J.; Tew, K.D. Novel role for glutathione Stransferase pi. Reg-ulator of protein S-Glutathionylation following oxidative and nitrosative stress. J. Biol. Chem. 2009, 284, 436–445, doi:10.1074/jbc.M805586200.
- Uemura, T.; Tsaprailis, G.; Gerner, E.W. GSTΠ stimulates caveolin-1-regulated polyamine uptake via actin remodeling. On-cotarget 2019, 10, 5713–5723, doi:10.18632/oncotarget.27192.
- Wetzelberger, K.; Baba, S.P.; Thirunavukkarasu, M.; Ho, Y.S.; Maulik, N.; Barski, O.A.; Conklin, D.J.; Bhatnagar, A. Postis-chemic deactivation of cardiac aldose reductase: Role of glutathione Stransferase P and glutaredoxin in regeneration of re-duced thiols from sulfenic acids. J. Biol. Chem. 2010, 285, 26135–26148, doi:10.1074/jbc.M110.146423.
- Zhang, J.; Ye, Z.W.; Chen, W.; Manevich, Y.; Mehrotra, S.; Ball, L.; Janssen-Heininger, Y.; Tew, K.D.; Townsend, D.M. S-Glutathionylation of estrogen receptor α affects dendritic cell function. J. Biol. Chem. 2018, 293, 4366–4380, doi:10.1074/jbc.M117.814327.
- Graminski, G.F.; Kubo, Y.; Armstrong, R.N. Spectroscopic and kinetic evidence for the thiolate anion of glutathione at the active site of glutathione S‐transferase. Biochem. (Mosc) 1989, 28, 3562–3568, doi:10.1021/bi00434a062.
- Deponte, M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim. Biophys. Acta 2013, 1830, 3217–3266, doi:10.1016/j.bbagen.2012.09.018.
- Ercolani, L.; Scire, A.; Galeazzi, R.; Massaccesi, L.; Cianfruglia, L.; Amici, A.; Piva, F.; Urbanelli, L.; Emiliani, C.; Principato, G.; et al. A possible S-glutathionylation of specific proteins by glyoxalase II: An in vitro and in silico study. Cell Biochem. Funct. 2016, 34, 620–627, doi:10.1002/cbf.3236.
- Burns, M.; Rizvi, S.H.M.; Tsukahara, Y.; Pimentel, D.R.; Luptak, I.; Hamburg, N.M.; Matsui, R.; Bachschmid, M.M. Role of Glutaredoxin-1 and Glutathionylation in Cardiovascular Diseases. Int. J. Mol. Sci. 2020, 21, 6803, doi:10.3390/ijms21186803.
- Xiao, Z.; La Fontaine, S.; Bush, A.I.; Wedd, A.G. Molecular Mechanisms of Glutaredoxin Enzymes: Versatile Hubs for Thi-ol-Disulfide Exchange between Protein Thiols and Glutathione. J. Mol. Biol. 2019, 431, 158–177, doi:10.1016/j.jmb.2018.12.006.
- Ren, X.; Zou, L.; Zhang, X.; Branco, V.; Wang, J.; Carvalho, C.; Holmgren, A.; Lu, J. Redox Signaling Mediated by Thiore-doxin and Glutathione Systems in the Central Nervous System. Antioxid. Redox Signal. 2017, 27, 989–1010, doi:10.1089/ars.2016.6925.
- Gallogly, M.M.; Starke, D.W.; Mieyal, J.J. Mechanistic and kinetic details of catalysis of thiol-disulfide exchange by glu-taredoxins and potential mechanisms of regulation. Antioxid. Redox Signal. 2009, 11, 1059–1081, doi:10.1089/ars.2008.2291.
- Beer, S.M.; Taylor, E.R.; Brown, S.E.; Dahm, C.C.; Costa, N.J.; Runswick, M.J.; Murphy, M.P. Glutaredoxin 2 catalyzes the re-versible oxidation and glutathionylation of mitochondrial membrane thiol proteins: Implications for mitochondrial redox regulation and antioxidant DEFENSE. J. Biol. Chem. 2004, 279, 47939–47951, doi:10.1074/jbc.M408011200.
- Ukuwela, A.A.; Bush, A.I.; Wedd, A.G.; Xiao, Z. Reduction Potentials of Protein Disulfides and Catalysis of Glutathionyla-tion and Deglutathionylation by Glutaredoxin Enzymes. Biochem. J. 2017, 474, 3799–3815, doi:10.1042/BCJ20170589.
- Zhang, J.; Ye, Z.W.; Chen, W.; Culpepper, J.; Jiang, H.; Ball, L.; Mehrotra, S.; Blumental-Perry, A.; Tew, K.D.; Townsend, D.M. Altered Redox Regulation and S-Glutathionylation of BiP Contribute to Bortezomib Resistance in Multiple Myeloma. Free Radic. Biol. Med. 2020, 160, 755–767, doi:10.1016/j.freeradbiomed.2020.09.013.
- Gallogly, M.M.; Mieyal, J.J. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr. Opin. Pharmacol. 2007, 7, 381–391, doi:10.1016/j.coph.2007.06.003.
- Muronetz, V.I.; Melnikova, A.K.; Saso, L.; Schmalhausen, E.V. Influence of oxidative stress on catalytic and non-glycolytic functions of glyceraldehyde-3-phosphate dehydrogenase. Curr. Med. Chem. 2020, 27, 2040–2058, doi:10.2174/0929867325666180530101057.
- Chalker, J.; Gardiner, D.; Kuksal, N.; Mailloux, R.J. Characterization of the impact of glutaredoxin-2 (GRX2) deficiency on superoxide/hydrogen peroxide release from cardiac and liver mitochondria. Redox Biol. 2018, 15, 216–227, doi:10.1016/j.redox.2017.12.006.
- Menon, D.; Board, P.G. A role for glutathione transferase Omega 1 (GSTO1-1) in the glutathionylation cycle. J. Biol. Chem. 2013, 288, 25769–25779, doi:10.1074/jbc.M113.487785.
- Board, P.G.; Menon, D. Structure, function and disease relevance of Omega-class glutathione transferases. Arch. Toxicol. 2016, 90, 1049–1067, doi:10.1007/s00204-016-1691-1.
- Hughes, M.M.; Hooftman, A.; Angiari, S.; Tummala, P.; Zaslona, Z.; Runtsch, M.C.; McGettrick, A.F.; Sutton, C.E.; Diskin, C.; Rooke, M.; et al. Glutathione Transferase Omega-1 Regulates NLRP3 Inflammasome Activation through NEK7 Deglutathi-onylation. Cell Rep. 2019, 29, 151–161.e5, doi:10.1016/j.celrep.2019.08.072.
- Park, J.W.; Mieyal, J.J.; Rhee, S.G.; Chock, P.B. Deglutathionylation of 2-Cys peroxiredoxin is specifically catalyzed by sul-firedoxin. J. Biol. Chem. 2009, 284, 23364–23374, doi:10.1074/jbc.M109.021394.
- Calderón, A.; Lázaro-Payo, A.; Iglesias-Baena, I.; Camejo, D.; Lázaro, J.J.; Sevilla, F.; Jiménez, A. Glutathionylation of Pea Chloroplast 2-Cys Prx and Mitochondrial Prx IIF Affects Their Structure and Peroxidase Activity and Sulfiredoxin Degluta-thionylates Only the 2-Cys Prx. Front. Plant Sci. 2017, 8, 118, doi:10.3389/fpls.2017.00118.
- Findlay, V.J.; Townsend, D.M.; Morris, T.E.; Fraser, J.P.; He, L.; Tew, K.D. A novel role for human sulfiredoxin in the rever-sal of glutathionylation. Cancer Res. 2006, 66, 6800–6806, doi:10.1158/0008-5472.CAN-06-0484.
- Bedhomme, M.; Adamo, M.; Marchand, C.H.; Couturier, J.; Rouhier, N.; Lemaire, S.D.; Zaffagnini, M.; Trost, P. Glutathi-onylation of cytosolic glyceraldehyde-3-phosphate dehydrogenase from the model plant Arabidopsis thaliana is reversed by both glutaredoxins and thioredoxins in vitro. Biochem. J. 2012, 445, 337–347, doi:10.1042/BJ20120505.
- Hanschmann, E.M.; Lonn, M.E.; Schutte, L.D.; Funke, M.; Godoy, J.R.; Eitner, S.; Hudemann, C.; Lillig, C.H. Both thioredoxin 2 and glutaredoxin 2 contribute to the reduction of the mitochondrial 2-Cys peroxiredoxin Prx3. J. Biol. Chem. 2010, 285, 40699–40705, doi:10.1074/jbc.M110.185827.
- Silva, G.M.; Netto, L.E.; Discola, K.F.; Piassa-Filho, G.M.; Pimenta, D.C.; Barcena, J.A.; Demasi, M. Role of glutaredoxin 2 and cytosolic thioredoxins in cysteinyl-based redox modification of the 20S proteasome. FEBS J. 2008, 275, 2942–2955, doi:10.1111/j.1742-4658.2008.06441.x.
- Subramani, J.; Kundumani-Sridharan, V.; Hilgers, R.H.; Owens, C.; Das, K.C. Thioredoxin Uses a GSH-independent Route to Deglutathionylate Endothelial Nitric-oxide Synthase and Protect against Myocardial Infarction. J. Biol. Chem. 2016, 291, 23374–23389, doi:10.1074/jbc.M116.745034.
- Peltoniemi, M.J.; Karala, A.R.; Jurvansuu, J.K.; Kinnula, V.L.; Ruddock, L.W. Insights into deglutathionylation reactions. Different intermediates in the glutaredoxin and protein disulfide isomerase catalyzed reactions are defined by the gam-ma-linkage present in glutathione. J. Biol. Chem. 2006, 281, 33107–33114, doi:10.1074/jbc.M605602200.
- Xie, Y.; Dahlin, J.L.; Oakley, A.J.; Casarotto, M.G.; Board, P.G.; Baell, J.B. Reviewing Hit Discovery Literature for Difficult Targets: Glutathione Transferase Omega-1 as an Example. J. Med. Chem. 2018, 61, 7448–7470, doi:10.1021/acs.jmedchem.8b00318.
- Lu, J.; Holmgren, A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014, 66, 75–87, doi:10.1016/j.freeradbiomed.2013.07.036.
- Subramani, J.; Kundumani-Sridharan, V.; Hilgers, R.H.; Owens, C.; Das, K.C. Thioredoxin Uses a GSH-independent Route to Deglutathionylate Endothelial Nitric-oxide Synthase and Protect against Myocardial Infarction. J. Biol. Chem. 2016, 291, 23374–23389, doi:10.1074/jbc.M116.745034.
- Peltoniemi, M.J.; Karala, A.R.; Jurvansuu, J.K.; Kinnula, V.L.; Ruddock, L.W. Insights into deglutathionylation reactions. Different intermediates in the glutaredoxin and protein disulfide isomerase catalyzed reactions are defined by the gam-ma-linkage present in glutathione. J. Biol. Chem. 2006, 281, 33107–33114, doi:10.1074/jbc.M605602200.
- Xie, Y.; Dahlin, J.L.; Oakley, A.J.; Casarotto, M.G.; Board, P.G.; Baell, J.B. Reviewing Hit Discovery Literature for Difficult Targets: Glutathione Transferase Omega-1 as an Example. J. Med. Chem. 2018, 61, 7448–7470, doi:10.1021/acs.jmedchem.8b00318.
- Lu, J.; Holmgren, A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014, 66, 75–87, doi:10.1016/j.freeradbiomed.2013.07.036.