
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Muhammad Amjad Nawaz | + 3411 word(s) | 3411 | 2021-06-02 08:27:52 | | | |
| 2 | Vivi Li | + 430 word(s) | 3841 | 2021-06-23 04:45:58 | | | | |
| 3 | Vivi Li | + 430 word(s) | 3841 | 2021-06-23 04:46:34 | | |
Video Upload Options
Domesticated crops suffer from major genetic bottlenecks while wild relatives retain higher genomic diversity. Wild soybean (Glycine soja Sieb. & Zucc.) is the presumed ancestor of cultivated soybean (Glycine max [L.] Merr.), and is an important genetic resource for soybean improvement. Among the East Asian habitats of wild soybean (China, Japan, Korea, and Northeastern Russia), the Korean peninsula is of great importance based on archaeological records, domestication history, and higher diversity of wild soybeans in the region. The collection and conservation of these wild soybean germplasms should be put on high priority. Chung’s Wild Legume Germplasm Collection maintains more than 10,000 legume accessions with an intensive and prioritized wild soybean germplasm collection (>6000 accessions) guided by the international code of conduct for plant germplasm collection and transfer. The center holds a library of unique wild soybean germplasms collected from East Asian wild habitats including the Korean mainland and nearby islands. The collection has revealed interesting and useful morphological, biochemical, and genetic diversity. This resource could be utilized efficiently in ongoing soybean improvement programs across the globe.
1. Introduction
2. Distribution and Conservation of Wild Soybean in Korea
2.1. Geographical Distribution of Wild Soybean in Korea
2.2. Archaeological Records and Background
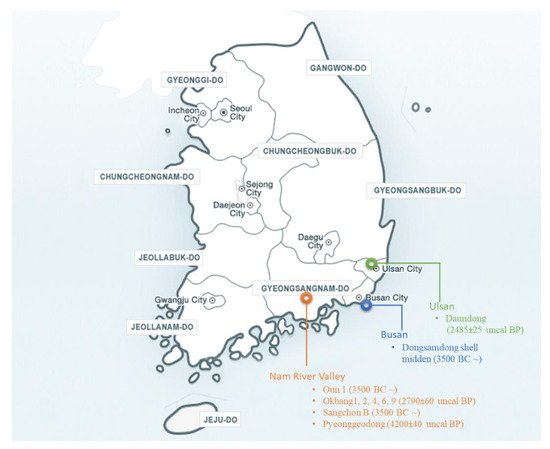
3. Collection of Wild Soybean Germplasm in Korea
| Instcode | Institute | No. of Accessions | % | WS | LR | BL | AC | OT |
|---|---|---|---|---|---|---|---|---|
| CHN001 | ICGR-CAAS | 32,021 | 14 | 21 | 79 | |||
| USA033 | SOY | 21,075 | 9 | 10 | 80 | 5 | 4 | 1 |
| KOR011 | RDAGB-GRD | 17,644 | 8 | <1 | 45 | 5 | 1 | 50 |
| TWN001 | AVRDC | 15,314 | 7 | <1 | <1 | 100 | ||
| BRA014 | CNPSO | 11,800 | 5 | 100 | ||||
| JPN003 | NIAS | 11,473 | 5 | 5 | 33 | 21 | 40 | |
| RUS001 | VIR | 6439 | 3 | 9 | 40 | 41 | 11 | |
| IND016 | AICRP-Soybean | 4022 | 2 | <1 | 100 | |||
| CIV005 | IDESSA | 3727 | 2 | 100 | ||||
| TWN006 | TARI | 2745 | 1 | 100 | ||||
| DEU146 | IPK | 2661 | 1 | 1 | 23 | 53 | 23 | |
| ZWE003 | CBICAU | 2236 | 1 | 100 | ||||
| IDN182 | ICRR | 2198 | 1 | <1 | 100 | |||
| AUS048 | ATCFC | 2121 | 1 | 3 | <1 | 38 | 52 | 6 |
| NGA039 | IITA | 1909 | 1 | 5 | 4 | 1 | 90 | |
| FRA060 | AMFO | 1582 | 1 | 100 | ||||
| THA005 | FCRI-DA/TH | 1510 | 1 | 100 | ||||
| MEX001 | INIA-Iguala | 1500 | 1 | 100 | ||||
| PHL130 | IPB-UPLB | 1381 | 1 | 100 | ||||
| UKR001 | IR | 1288 | 1 | 3 | 1 | 21 | 72 | 3 |
| COL017 | ICA/REGION 1 | 1235 | 1 | <1 | 64 | 13 | 22 | |
| SRB002 | IFVCNS | 1200 | 1 | 100 | ||||
| ROM002 | ICCPT Fundul | 1024 | <1 | 62 | 38 | <1 | ||
| Others (166) | 81,839 | 36 | 11 | 4 | 27 | 51 | ||
| Total | 229,944 | 100 | 6 | 17 | 7 | 13 | 56 | |
| ICGR-CAAS | Institute of Crop Germplasm Resources, Chinese Academy of Agricultural Sciences | |||||||
| SOY | Soybean Germplasm Collection, United States Department of Agriculture, Agricultural Research Services | |||||||
| RDAGB-GRD | Genetic Resources Division, National Institute of Agricultural Biotechnology, Rural Development Administration (Korea) | |||||||
| AVRDC | World Vegetable Centre (former Asian Vegetable Research and Development Centre) | |||||||
| CNPSO | Embrapa Soja (Brazil) | |||||||
| NIAS | National Institute of Agrobiological Sciences (Japan) | |||||||
| VIR | N.I. Vavilov All-Russian Scientific Research Institute of Plant Industry (Russian Federation) | |||||||
| AICRP-Soybean | All India Coordinated Research Project on Soybean (India) | |||||||
| IDESSA | Institut des Savanes (Côte d’Ivoire) | |||||||
| TARI | Taiwan Agricultural Research Institute | |||||||
| IPK | Genebank, Leibniz Institute of Plant Genetics and Crop Plant Research (Germany) | |||||||
| CBICAU | Crop Breeding Institute (Zimbabwe) | |||||||
| ICRR | Indonesian Centre for Rice Research | |||||||
| ATCFC | Australian Tropical Crops & Forages Genetic Resources Centre | |||||||
| IITA | International Institute of Tropical Agriculture | |||||||
| AMFO | G.I.E. Amelioration Fourragère (France) | |||||||
| FCRI-DA/TH | Field Crops Research Institute–Department of Agriculture (Thailand) | |||||||
| INIA-Iguala | Estación de Iguala, Instituto Nacional de Investigaciones Agrícolas (Mexico) | |||||||
| IPB-UPLB | Institute of Plant Breeding, College of Agriculture, University of the Philippines, Los Baños College (Philippines) | |||||||
| IR | Institute of Plant Production n.a. V.Y. Yurjev of UAAS (Ukraine) | |||||||
| ICA/REGION 1 | Corporación Colombiana de Investigación Agropecuaria Tibaitata (Colombia) | |||||||
| IFVCNS | Institute for Field and Vegetable Crops (Serbia) | |||||||
| ICCPT Fundul | Research Institute for Cereals and Technical Plants Fundulea (Romania) | |||||||
| Species | No. of Accessions |
|---|---|
| Glycine max (L.) Merr | 600 |
| Glycine soja | 6232 |
| Amphicarpaea edgeworthii | 1300 |
| Vigna vexillata | 810 |
| Rhynchosia volubilis | 225 |
| Phaseolus nipponensis | 700 |
| Other wild legumes Dunbaria villosa (Thunb.) Makino Vigna umbellate (Thumb.) Ohwi & Ohashi Vigna angularis var. nipponensis (ohwi) ohwi & ohashi Vigna angularis (Willd.) Ohwi & Ohashi |
700 |
| Total | 10,567 |
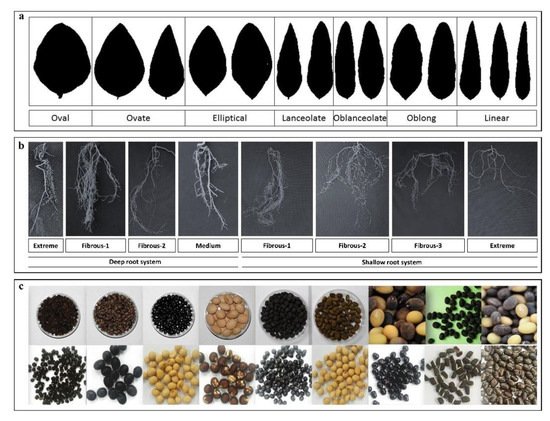
3.1. Chung’s Wild Legume Germplasm Collection
3.2. In situ Conservation of G. soja at CWLGC

| Trait | Gene/QTL | Chromosome/Locus Group | Source (accession) | Reference |
|---|---|---|---|---|
| Yield Related Traits | ||||
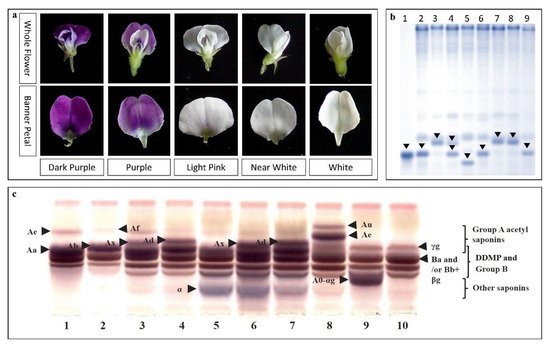
| Flower color | Dihydroflavonol-4-reductase 4 (w4-s1 allele) (DFR) | B2 (Ch 14) | WS (CW13133) | [46] |
| Flavonoid 3′5′-hydroxylase (F3′5′H) and DFR | B2 (Ch 14) | WS (CW12700 and CW13381) | [45] | |
| DFR2 (w4-s1) | B2 (Ch 14) | WS (CW13133) × CS (IT182932) | [47] | |
| Oil and local breeding related traits | QTLs | 3, 8, 13, 17, | WS × CS | [17] |
| Shoot fresh weight | QTLs | 6, 15, 19 | WS (PI 483463) × CS (Hutcheson) | [48] |
| Biological nitrogen fixation related traits | QTLs | 7,8,12,17,18 | WS (W05) × CS (C08) | [49] |
| Small seed, Yellow seed color, Soy sprout |
WS (PI135624) | [50] | ||
| High yield | B2 (U26) | WS (PI 407305) | [51] | |
| Yield, height and maturity | QTLs | C2, E, K and M | WS × CS | [52] |
| Seed yield, 100-seed weight, seed filling period, maturity, height, lodging | QTLs | A1, J, N, H, F, L | WS × CS | [53] |
| QTLs | Multiple | CS (NN86-4) × WS (PI342618B) | [54] | |
| QTLs | 1, 2, 6, 8, 13, 14, 17, 20 | CS (Williams 82) × WS (PI 366121) | [55] | |
| Biotic Stress Tolerance | ||||
| Soybean cyst nematode resistance | Rhg1, Rhg4 locus and QTLs | G, A2, B1 | CS (Magellan) × WS (PI 404198A) | [56] |
| Glyma18g196200, Glyma18g077900, Glyma18g078000, Glyma18g106800, Glyma18g107000, Glyma18g107100 | Ch18 | WS | [57] | |
| Glyma18g106800, Glyma18g064100 | ||||
| QTLs (cqSCN-006, cqSCN-007) | I, J, K, O | WS (PI 468916, 464925B) | [58][59] | |
| Markers (A245_1 and Satt598) | 15, 18 | WS (PI 468916) | [60] | |
| Resistance to southern root knot nematode | WS | [61] | ||
| Foxglove aphid resistance (Aulacorthum solani) |
Raso2 | 7 | CS (Williams 82) × CS (PI 366121) | [62] |
| Aphid resistance (Aphis glycines) | Rag3c and Rag6 | 8, 16 | WS (85-32) | [63] |
| QTLs | 8, 16, | WS WS (PI 518282) |
[64] [65] |
|
| Sclerotinia stem rot | QTLs | 3, 8, 20 | WS × CS | [66][67][68] |
| Resistance to Mung bean yellow mosaic India virus | Ch 8 and 14 | WS (PI 393551) | [69] | |
| Resistance to Phtophthora soja | QTLs | J, I, G (Ch 16, 18, 20) | WS (PI 407162) × CS (V71-370) | [70] |
| Abiotic Stress Tolerance | ||||
| Salt tolerance | WS (N23232 BB52) | [71] | ||
| Allele (Ncl locus) | WS (PI 483463) | [72] | ||
| QTLs | D2 (Ch 17) | WS (JWS156-1) | [73] | |
| QTLs | 3 | WS | [74] | |
| RLKs CaMs, JA/SA Signaling genes, MAPKs, WRKYs | [67][68] | |||
| Tolerance to herbicide metribuzin | WS (PI 245331, PI 163453) | [75] | ||
| Root traits | QTLs | 8, 12 | WS (PI 326582A) × CS (PI 552538) | [76] |
| Root architecture | Glyma15g42220, Glyma06g46210, Glyma06g45910, Glyma06g45920, Glyma07g32480 | 6, 7, 15 | WS (PI 407162) × CS (V71-370) | [77] |
| Alkalinity tolerance | ALMT, LEA, ABC transporter, GLR, NRT/POT and SLAH genes | Multiple | WS (N24852) | [78] |
| Drought tolerance | GsWRKY20 | [79] | ||
| Nutrition | ||||
| Seed protein content | Marker pA-245 | C | CS (A81-356022) × WS (PI 468916) | [80] |
| Seed saturated fatty acid contents | SNPs | GS | [81] | |
| Glyma14g121400, Glyma16g068500 | 14, 16 | |||
| Seed unsaturated fatty acids | Glyma16g014000, Glyma07g112100 | 7, 16 | ||
| Linolenic acid | QTLs | Multiple | WS × CS | [82] |
| Pearl Japanese fermented product (Natto) | WS | [61] | ||
| Protein, oil, palmitic acid, stearic acid, oleic acid, linoleic acid, lenolenic acid | QTLs | 2, 7, 14, 16 | WS | [81] |
| Saponin A | Sg-1 | Ch 7 | WS | [65][83] |
| Sg-5 (Glyma15g39090) | Ch 15 | WS × CS | [84][69] | |
| Glyma15g39090 | Ch 15 | WS | [36] | |
| Seed antioxidant, phenolics, and flavonoids | GmMATE1,2,4 | Ch 18,19 | WS (W05) × CS (C08) | [85] |
References
- Kofsky, J.; Zhang, H.; Song, B.-H. The untapped genetic reservoir: the past, current, and future applications of the wild soybean (Glycine soja). Front. Plant Sci. 2018, 9.
- Nawaz, M.A.; Rehman, H.M.; Imtiaz, M.; Baloch, F.S.; Lee, J.D.; Yang, S.H.; Lee, S.I.; Chung, G. Systems Identification and Characterization of Cell Wall Reassembly and Degradation Related Genes in Glycine max (L.) Merill, a Bioenergy Legume. Sci. Rep. 2017, 7, 10862.
- Lam, H.-M.; Remais, J.; Fung, M.-C.; Xu, L.; Sun, S.S.-M. Food supply and food safety issues in China. The Lancet 2013, 381, 2044–2053.
- Considine, M.J.; Siddique, K.H.; Foyer, C.H. Nature’s pulse power: Legumes, food security and climate change. J. Exp. Bot. 2017, 68, 1815–1818.
- Allender, C. The Second Report on the State of the World’s Plant. Genetic Resources for Food and Agriculture; FAO Commission on Genetic Resources for Food and Agriculture: Rome, Italy, 2010.
- Lin, B.B. Resilience in agriculture through crop diversification: adaptive management for environmental change. BioScience 2011, 61, 183–193.
- Hyten, D.L.; Song, Q.; Zhu, Y.; Choi, I.-Y.; Nelson, R.L.; Costa, J.M.; Specht, J.E.; Shoemaker, R.C.; Cregan, P.B. Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci. USA 2006, 103, 16666–16671.
- Cowling, W.; Li, L.; Siddique, K.; Henryon, M.; Berg, P.; Banks, R.; Kinghorn, B. Evolving gene banks: Improving diverse populations of crop and exotic germplasm with optimal contribution selection. J. Exp. Bot. 2016, 68, 1927–1939.
- Foyer, C.H.; Lam, H.-M.; Nguyen, H.T.; Siddique, K.H.; Varshney, R.K.; Colmer, T.D.; Cowling, W.; Bramley, H.; Mori, T.A.; Hodgson, J.M. Neglecting legumes has compromised human health and sustainable food production. Nat. Plant. 2016, 2, 16112.
- Muñoz, N.; Liu, A.; Kan, L.; Li, M.-W.; Lam, H.-M. Potential uses of wild germplasms of grain legumes for crop improvement. Int. J. Mol. Sci. 2017, 18, 328.
- Chung, G.; Singh, R.J. Broadening the genetic base of soybean: A multidisciplinary approach. Crit. Rev. Plant Sci. 2008, 27, 295–341.
- Ladizinsky, G.; Newell, C.; Hymowitz, T. Wide crosses in soybeans: Prospects and limitations. Euphytica 1979, 28, 421–423.
- Singh, R.; Nelson, R.L. Methodology for creating alloplasmic soybean lines by using Glycine tomentella as a maternal parent. Plant Breed. 2014, 133, 624–631.
- Singh, R.J. Cytogenetics and genetic introgression from wild relatives in soybean. Nucleus 2019, 62, 3–14.
- Singh, R.; Hymowitz, T. The genomic relationship between Glycine max (L.) Merr. and G. soja Sieb. and Zucc. as revealed by pachytene chromosome analysis. Theor. Appl. Genet. 1988, 76, 705–711.
- He, S.-L.; Wang, Y.-S.; Li, D.-Z.; Yi, T.-S. Environmental and historical determinants of patterns of genetic differentiation in wild soybean (Glycine soja Sieb. et Zucc). Sci. Rep. 2016, 6, 22795.
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408–414.
- Wang, L.-x.; Lin, F.-y.; LI, L.-h.; Wei, L.; Zhe, Y.; LUAN, W.-j.; PIAO, R.-h.; Yuan, G.; NING, X.-c.; Li, Z. Genetic diversity center of cultivated soybean (Glycine max) in China–New insight and evidence for the diversity center of Chinese cultivated soybean. J. Integr. Agric. 2016, 15, 2481–2487.
- Leamy, L.J.; Lee, C.R.; Song, Q.; Mujacic, I.; Luo, Y.; Chen, C.Y.; Li, C.; Kjemtrup, S.; Song, B.H. Environmental versus geographical effects on genomic variation in wild soybean (Glycine soja) across its native range in northeast Asia. Ecol. Evolut. 2016, 6, 6332–6344.
- Hay, F.R.; Probert, R.J. Advances in seed conservation of wild plant species: A review of recent research. Conserv. Physiol. 2013, 1.
- Castañeda-Álvarez, N.P.; Khoury, C.K.; Achicanoy, H.A.; Bernau, V.; Dempewolf, H.; Eastwood, R.J.; Guarino, L.; Harker, R.H.; Jarvis, A.; Struik, P.C.; et al. Global conservation priorities for crop wild relatives. Nat. Plant. 2016, 2, 16022.
- Lee, J.D.; Shannon, J.; Vuong, T.; Moon, H.; Nguyen, H.; Tsukamoto, C.; Chung, G. Genetic diversity in wild soybean (Glycine soja Sieb. and Zucc.) accessions from southern islands of Korean peninsula. Plant Breed. 2010, 129, 257–263.
- Nawaz, M.A.; Yang, S.H.; Rehman, H.M.; Baloch, F.S.; Lee, J.D.; Park, J.H.; Chung, G. Genetic diversity and population structure of Korean wild soybean (Glycine soja Sieb. and Zucc.) inferred from microsatellite markers. Biochem. Syst. Ecol. 2017, 71, 87–96.
- Carter, T.; Nelson, R.; Sneller, C.; Cui, Z. Genetic diversity in soybean. Soybean Monogr. Am. Soc. Agron. Madison Wis. USA 2004.
- Li, M.-W.; Wang, Z.; Jiang, B.; Kaga, A.; Wong, F.-L.; Zhang, G.; Han, T.; Chung, G.; Nguyen, H.; Lam, H.-M. Impacts of genomic research on soybean improvement in East Asia. Theor. Appl. Genet. 2019, 1–24.
- Lee, G.-A.; Crawford, G.W.; Liu, L.; Sasaki, Y.; Chen, X. Archaeological soybean (Glycine max) in East Asia: Does size matter? PLoS ONE 2011, 6, e26720.
- Crawford, G.W.; Lee, G.-A. Agricultural origins in the Korean Peninsula. Antiquity 2003, 77, 87.
- Barnes, G.L. Archaeology of East. Asia: The Rise of Civilization in China, Korea and Japan; Oxbow Books: Oxford, UK; Casemate Publishing: Pennsylvania, PA, USA, 2015.
- Maxted, N.; Kell, S. Establishment of a global network for the in situ conservation of crop wild relatives: Status and needs. FAO Comm. Genet. Resour. Food Agric. Rome 2009, 266, 509.
- Kim, M.Y.; Van, K.; Kang, Y.J.; Kim, K.H.; Lee, S.-H. Tracing soybean domestication history: From nucleotide to genome. Breed. Sci. 2012, 61, 445–452.
- Lee, S.; Kim, J.-H.; Sundaramoorthy, J.; Park, G.T.; Lee, J.-D.; Kim, J.H.; Chung, G.; Seo, H.S.; Song, J.T. Identification of GmSALT3 haplotypes and development of molecular markers based on their diversity associated with salt tolerance in soybean. Mol. Breed. 2018, 38, 86.
- Panneerselvam, K.; Tsukamoto, C.; Honda, N.; Kikuchi, A.; Lee, J.D.; Yang, S.H.; Chung, G. Saponin polymorphism in the Korean wild soybean (Glycine soja Sieb. and Zucc.). Plant Breed. 2013, 132, 121–126.
- Krishnamurthy, P.; Lee, J.M.; Tsukamoto, C.; Takahashi, Y.; Singh, R.J.; Lee, J.D.; Chung, G. Evaluation of genetic structure of Korean wild soybean (Glycine soja) based on saponin allele polymorphism. Genet. Res. Crop Evol. 2014, 61, 1121–1130.
- Krishnamurthy, P.; Tsukamoto, C.; Singh, R.J.; Lee, J.-D.; Kim, H.-S.; Yang, S.-H.; Chung, G. The Sg-6 saponins, new components in wild soybean (Glycine soja Sieb. and Zucc.): polymorphism, geographical distribution and inheritance. Euphytica 2014, 198, 413–424.
- Krishnamurthy, P.; Tsukamoto, C.; Takahashi, Y.; Hongo, Y.; Singh, R.J.; Lee, J.D.; Chung, G. Comparison of saponin composition and content in wild soybean (Glycine soja Sieb. and Zucc.) before and after germination. Biosci. Biotechnol. Biochem. 2014, 78, 1988–1996.
- Rehman, H.M.; Nawaz, M.A.; Shah, Z.H.; Yang, S.H.; Chung, G. Functional characterization of naturally occurring wild soybean mutant (sg-5) lacking astringent saponins using whole genome sequencing approach. Plant Sci. 2017.
- Sundaramoorthy, J.; Park, G.T.; Mukaiyama, K.; Tsukamoto, C.; Chang, J.H.; Lee, J.-D.; Kim, J.H.; Seo, H.S.; Song, J.T. Molecular elucidation of a new allelic variation at the Sg-5 gene associated with the absence of group A saponins in wild soybean. PLoS ONE 2018, 13, e0192150.
- Krishnamurthy, P.; Lee, J.D.; Ha, B.K.; Chae, J.H.; Song, J.T.; Tsukamoto, C.; Singh, R.J.; Chung, G. Genetic characterization of group A acetylsaponin-deficient mutants from wild soybean (Glycine soja Sieb. and Zucc.). Plant Breed. 2015, 134, 316–321.
- Rehman, H.M.; Nawaz, M.A.; Shah, Z.H.; Chung, G.; Yang, S.H. Molecular Elucidation of Two Novel Seed Specific Flavonoid Glycosyltransferases in Soybean. J. Plant Biol. 2018, 61, 320–329.
- Krishnamurthy, P.; Singh, R.J.; Tsukamoto, C.; Park, J.H.; Lee, J.D.; Chung, G. Kunitz trypsin inhibitor polymorphism in the Korean wild soybean (Glycine soja Sieb. & Zucc.). Plant Breed. 2013, 132, 311–316.
- Nawaz, M.A.; Golokhvast, K.S.; Rehman, H.M.; Tsukamoto, C.; Kim, H.-S.; Yang, S.H.; Chung, G. Soyisoflavone diversity in wild soybeans (Glycine soja Sieb. & Zucc.) from the main centres of diversity. Biochem. Syst. Ecol. 2018, 77, 16–21.
- Tsukamoto, C.; Nawaz, M.A.; Kurosaka, A.; Le, B.; Lee, J.D.; Son, E.; Yang, S.H.; Kurt, C.; BALOCH, F.S.; Chung, G. Isoflavone profile diversity in Korean wild soybeans (Glycine soja Sieb. & Zucc.). Turk. J. Agric. For. 2018, 42, 248–261.
- Chae, J.-H.; Ha, B.-K.; Chung, G.; Park, J.-E.; Park, E.; Ko, J.-M.; Shannon, J.G.; Song, J.T.; Lee, J.-D. Identification of environmentally stable wild soybean genotypes with high alpha-linolenic acid concentration. Crop Sci. 2015, 55, 1629–1636.
- Ali, M.; Krishnamurthy, P.; El-Hadary, M.; Kim, J.; Nawaz, M.; Yang, S.; Chung, G. Identification and expression profiling of a new β-amyrin synthase gene (GmBAS3) from soybean. Russ. J. Plant physiol. 2016, 63, 383–390.
- Sundaramoorthy, J.; Park, G.T.; Chang, J.H.; Lee, J.-D.; Kim, J.H.; Seo, H.S.; Chung, G.; Song, J.T. Identification and molecular analysis of four new alleles at the W1 locus associated with flower color in soybean. PLoS ONE 2016, 11, e0159865.
- Park, G.T.; Sundaramoorthy, J.; Lee, S.; Lee, J.-D.; Kim, J.H.; Park, S.-K.; Seo, H.S.; Chung, G.; Song, J.T. Color Variation in a Novel Glycine soja Mutant W4-S1 with Pinkish-White Flowers Is Controlled by a Single Recessive Allele at the W4 Locus. Crop Sci. 2017, 57, 3112–3121.
- Park, G.T.; Sundaramoorthy, J.; Lee, S.; Lee, J.-D.; Kim, J.H.; Park, S.-K.; Seo, H.S.; Chung, G.; Song, J.T. Color Variation in a Novel Mutant with Pinkish-White Flowers Is Controlled by a Single Recessive Allele at the Locus. Crop Sci. 2017, 57, 3112–3121.
- Asekova, S.; Kulkarni, K.P.; Patil, G.; Kim, M.; Song, J.T.; Nguyen, H.T.; Shannon, J.G.; Lee, J.-D. Genetic analysis of shoot fresh weight in a cross of wild (G. soja). Mol. Breed. 2016, 36, 1–15.
- Muñoz, N.; Qi, X.; Li, M.; Xie, M.; Gao, Y.; Cheung, M.; Wong, F.; Lam, H. Improvement in nitrogen fixation capacity could be part of the domestication process in soybean. Heredity 2016, 117, 84–93.
- Fehr, W.; Cianzio, S.; Welke, G. Registration of’SS202’soybean. Crop Sci. 1990, 30.
- Concibido, V.; La Vallee, B.; Mclaird, P.; Pineda, N.; Meyer, J.; Hummel, L.; Yang, J.; Wu, K.; Delannay, X. Introgression of a quantitative trait locus for yield from Glycine soja into commercial soybean cultivars. Theor. Appl. Genet. 2003, 106, 575–582.
- Wang, D.; Graef, G.; Procopiuk, A.; Diers, B. Identification of putative QTL that underlie yield in interspecific soybean backcross populations. Theor. Appl. Genet. 2004, 108, 458–467.
- Li, D.; Pfeiffer, T.; Cornelius, P. Soybean QTL for yield and yield components associated with alleles. Crop Sci. 2008, 48, 571–581.
- Wang, W.; Li, X.; Chen, S.; Song, S.; Gai, J.; Zhao, T. Using presence/absence variation markers to identify the QTL/allele system that confers the small seed trait in wild soybean (Glycine soja Sieb. & Zucc.). Euphytica 2016, 208, 101–111.
- Kulkarni, K.P.; Asekova, S.; Lee, D.-H.; Bilyeu, K.; Song, J.T.; Lee, J.-D. Mapping QTLs for 100-seed weight in an interspecific soybean cross of Williams 82 (Glycine max) and PI 366121 (Glycine soja). Crop Pasture Sci. 2017, 68, 148–155.
- Guo, B.; Sleper, D.A.; Nguyen, H.T.; Arelli, P.R.; Shannon, J.G. Quantitative trait loci underlying resistance to three soybean cyst nematode populations in soybean PI 404198A. Crop Sci. 2006, 46, 224–233.
- Zhang, H.; Li, C.; Davis, E.L.; Wang, J.; Griffin, J.D.; Kofsky, J.; Song, B.-H. Genome-wide association study of resistance to soybean cyst nematode (Heterodera glycines) HG Type 2.5. 7 in wild soybean (Glycine soja). Front. Plant Sci. 2016, 7, 1214.
- Winter, S.M.; Shelp, B.J.; Anderson, T.R.; Welacky, T.W.; Rajcan, I. QTL associated with horizontal resistance to soybean cyst nematode in Glycine soja PI464925B. Theor. Appl. Genet. 2007, 114, 461–472.
- Yu, N.; Diers, B.W. Fine mapping of the SCN resistance QTL cqSCN-006 and cqSCN-007 from Glycine soja PI 468916. Euphytica 2017, 213, 54.
- Wang, D.; Diers, B.W.; Arelli, P.; Shoemaker, R. Loci underlying resistance to race 3 of soybean cyst nematode in Glycine soja plant introduction 468916. Theor. Appl. Genet. 2001, 103, 561–566.
- Carter, T.E.; Huei, E.; Burton, J.; Farmer, F.; Gizlice, Z. Registration of ‘Pearl’soybean. Crop Sci. 1995, 35, 1713.
- Lee, J.S.; Yoo, M.-h.; Jung, J.K.; Bilyeu, K.D.; Lee, J.-D.; Kang, S. Detection of novel QTLs for foxglove aphid resistance in soybean. Theor. Appl. Genet. 2015, 128, 1481–1488.
- Zhang, S.; Zhang, Z.; Wen, Z.; Gu, C.; An, Y.-Q.C.; Bales, C.; DiFonzo, C.; Song, Q.; Wang, D. Fine mapping of the soybean aphid-resistance genes Rag6 and Rag3c from Glycine soja 85-32. Theor. Appl. Genet. 2017, 130, 2601–2615.
- Zhang, S.; Zhang, Z.; Bales, C.; Gu, C.; DiFonzo, C.; Li, M.; Song, Q.; Cregan, P.; Yang, Z.; Wang, D. Mapping novel aphid resistance QTL from wild soybean, Glycine soja 85-32. Theor. Appl. Genet. 2017, 130, 1941–1952.
- Hill, C.B.; Li, Y.; Hartman, G.L. Resistance of Glycine species and various cultivated legumes to the soybean aphid (Homoptera: Aphididae). J. Econ. Entomol. 2004, 97, 1071–1077.
- Iquira, E.; Humira, S.; François, B. Association mapping of QTLs for sclerotinia stem rot resistance in a collection of soybean plant introductions using a genotyping by sequencing (GBS) approach. BMC Plant Biol. 2015, 15, 5.
- Zhang, H.; Song, B.-H. RNA-seq data comparisons of wild soybean genotypes in response to soybean cyst nematode (Heterodera glycines). Genomics Data 2017, 14, 36–39.
- Zhang, H.; Song, Q.; Griffin, J.D.; Song, B.-H. Genetic architecture of wild soybean (Glycine soja) response to soybean cyst nematode (Heterodera glycines). Mol. Genet. Genomics 2017, 292, 1257–1265.
- Rani, A.; Kumar, V.; Gill, B.; Shukla, S.; Rathi, P.; Singh, R. Mapping of duplicate dominant genes for Mungbean yellow mosaic India virus resistance in Glycine soja. Crop Sci. 2018, 58, 1566–1574.
- Tucker, D.; Maroof, S.; Mideros, S.; Skoneczka, J.; Nabati, D.; Buss, G.; Hoeschele, I.; Tyler, B.; St Martin, S.; Dorrance, A. Mapping quantitative trait loci for partial resistance to Phytophthora sojae in a soybean interspecific cross. Crop Sci. 2010, 50, 628–635.
- Luo, Q.; Yu, B.; Liu, Y. Differential sensitivity to chloride and sodium ions in seedlings of Glycine max and G. soja under NaCl stress. J. Plant Physiol. 2005, 162, 1003–1012.
- Lee, J.-D.; Shannon, J.G.; Vuong, T.D.; Nguyen, H.T. Inheritance of salt tolerance in wild soybean (Glycine soja Sieb. and Zucc.) accession PI483463. J. Hered. 2009, 100, 798–801.
- Tuyen, D.; Lal, S.; Xu, D. Identification of a major QTL allele from wild soybean (Glycine soja Sieb. & Zucc.) for increasing alkaline salt tolerance in soybean. Theor. Appl. Genet. 2010, 121, 229–236.
- Ha, B.-K.; Vuong, T.D.; Velusamy, V.; Nguyen, H.T.; Shannon, J.G.; Lee, J.-D. Genetic mapping of quantitative trait loci conditioning salt tolerance in wild soybean (Glycine soja) PI 483463. Euphytica 2013, 193, 79–88.
- Kilen, T.C.; He, G. Identification and inheritance of metribuzin tolerance in wild soybean. Crop Sci. 1992, 32, 684–685.
- Manavalan, L.P.; Prince, S.J.; Musket, T.A.; Chaky, J.; Deshmukh, R.; Vuong, T.D.; Song, L.; Cregan, P.B.; Nelson, J.C.; Shannon, J.G. Identification of novel QTL governing root architectural traits in an interspecific soybean population. PLoS ONE 2015, 10, e0120490.
- Prince, S.J.; Song, L.; Qiu, D.; dos Santos, J.V.M.; Chai, C.; Joshi, T.; Patil, G.; Valliyodan, B.; Vuong, T.D.; Murphy, M. Genetic variants in root architecture-related genes in a Glycine soja accession, a potential resource to improve cultivated soybean. BMC Genomics 2015, 16, 132.
- Zhang, J.; Wang, J.; Jiang, W.; Liu, J.; Yang, S.; Gai, J.; Li, Y. Identification and analysis of NaHCO3 stress responsive genes in wild soybean (Glycine soja) roots by RNA-seq. Front. Plant Sci. 2016, 7, 1842.
- Ning, W.; Zhai, H.; Yu, J.; Liang, S.; Yang, X.; Xing, X.; Huo, J.; Pang, T.; Yang, Y.; Bai, X. Overexpression of Glycine soja WRKY20 enhances drought tolerance and improves plant yields under drought stress in transgenic soybean. Mol. Breed. 2017, 37, 19.
- Diers, B.W.; Keim, P.; Fehr, W.; Shoemaker, R. RFLP analysis of soybean seed protein and oil content. Theor. Appl. Genet. 1992, 83, 608–612.
- Leamy, L.J.; Zhang, H.; Li, C.; Chen, C.Y.; Song, B.-H. A genome-wide association study of seed composition traits in wild soybean (Glycine soja). BMC Genomics 2017, 18, 18.
- Pantalone, V.; Rebetzke, G.; Burton, J.; Wilson, R. Genetic regulation of linolenic acid concentration in wild soybean Glycine soja accessions. J. Am. Oil Chem. Soc. 1997, 74, 159–163.
- Park, J.; Kim, J.H.; Krishnamurthy, P.; Tsukamoto, C.; Song, J.T.; Chung, G.; Shannon, J.G.; Lee, J.-D. Characterization of a New Allele of the Saponin-Synthesizing Gene in Soybean. Crop Sci. 2016, 56, 385–391.
- Yano, R.; Takagi, K.; Takada, Y.; Mukaiyama, K.; Tsukamoto, C.; Sayama, T.; Kaga, A.; Anai, T.; Sawai, S.; Ohyama, K. Metabolic switching of astringent and beneficial triterpenoid saponins in soybean is achieved by a loss-of-function mutation in cytochrome P450 72A69. Plant J. 2017, 89, 527–539.
- Li, M.-W.; Muñoz, N.B.; Wong, C.-F.; Wong, F.-L.; Wong, K.-S.; Wong, J.W.-H.; Qi, X.; Li, K.-P.; Ng, M.-S.; Lam, H.-M. QTLs regulating the contents of antioxidants, phenolics, and flavonoids in soybean seeds share a common genomic region. Front. Plant Sci. 2016, 7.
- Fang, C.; Ma, Y.; Yuan, L.; Wang, Z.; Yang, R.; Zhou, Z.; Liu, T.; Tian, Z. Chloroplast DNA Underwent Independent Selection from Nuclear Genes during Soybean Domestication and Improvement. J. Genet. Genomics = Yi Chuan Xue Bao 2016, 43, 217.
- Asaf, S.; Khan, A.L.; Khan, M.A.; Imran, Q.M.; Kang, S.-M.; Al-Hosni, K.; Jeong, E.J.; Lee, K.E.; Lee, I.-J. Comparative analysis of complete plastid genomes from wild soybean (Glycine soja) and nine other Glycine species. PLoS ONE 2017, 12, e0182281.




