
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Monica Ballarino | + 2897 word(s) | 2897 | 2021-09-29 10:01:24 | | | |
| 2 | Lindsay Dong | + 122 word(s) | 3019 | 2021-09-30 07:47:43 | | |
Video Upload Options
In skeletal muscle, regeneration is driven by a reservoir of resident progenitors, called satellite cells, able to efficiently replenish damaged muscle [44]. These cells are not present in the adult cardiac muscle, although a regenerative response, mediated by the proliferation of pre-existing cardiomyocytes, occurs in mice during the first week of life [45,46,47]. Temporal and tissue-specific nuances in the process of regeneration may underlie the participation of still unknown protagonists, whose ability to fine-tune myogenic expression becomes critical in both physiological and pathological conditions. The peculiar properties of RNA, along with its tissue specificity, satisfy the requirements for its integration in regenerative networks and will surely pave the way for future applications in medicine.
1. Introduction
2. Applications of Small and Long RNAs as Therapeutic Tools for Muscle Regeneration
2.1. Small Non-Coding RNAs
2.1.1. microRNA (miRNA)
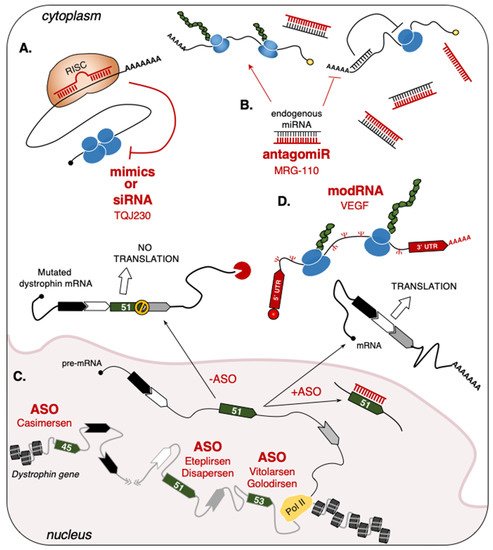
2.1.2. Single-Stranded Antisense Oligonucleotides (ASO)
| Drug | RNA Type | Target | Disease/Condition | Company | Phase | Reference |
|---|---|---|---|---|---|---|
| MRG-110 | Anti-miR | miR-92a | Wound Healing | miRagen (Viridian) |
Phase I | NCT03603431 |
| Spinraza (Nusinersen) |
ASO | SMN2 | SMA | Ionis | FDA/EMA approved |
NDA:209531 EMEA/H/C/004312 |
| Eteplirsen (Exondys 51) |
ASO | Dystrophin | DMD | Sarepta Inotersen |
FDA approved |
NDA:206488 |
| Drisapersen (Kyndrisa) |
ASO | Dystrophin | DMD | BioMarin | Phase III | NCT02636686 |
| Vitolarsen (Viltepso) |
ASO | Dystrophin | DMD | Nippon Shinyaku | FDA approved |
NDA:212154 |
| Golodirsen (Vyondis 53) |
ASO | Dystrophin | DMD | Sarepta Therapeutics |
FDA approved |
NDA:211970 |
| Casimersen (Amondys 45) |
ASO | Dystrophin | DMD | Sarepta Therapeutics |
FDA approved |
NDA:213026 |
| TQJ230 | siRNA | Apo(a) | Cardiovascular Disease, Elevated Lp(a) |
Novartis | Phase III | NCT04023552 |
| AZD8601 | mRNA | VEGF | Ischemic Heart Disease | Moderna, Astrazeneca |
Phase II | NCT03370887 |
| HEARTBiT | miR | Biomarker | Heart Transplant Rejection |
NCT03575910 | ||
| CRUCIAL | Circulating RNAs | Biomarker | Acute Heart Failure | NCT03345446 |
2.1.3. Short-Interfering RNA (siRNA)
2.2. Long-Sized RNAs
2.2.1. Protein-Coding RNAs
2.2.2. Non-Coding RNAs
Cytoplasmic ncRNAs
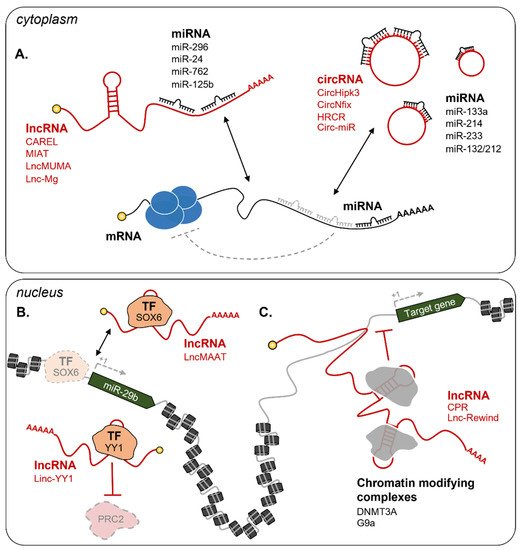
Nuclear ncRNAs
3. RNA as a Diagnostic Molecule for Muscle Diseases
4. Conclusions and Perspectives
References
- Brenner, S.; Jacob, F.; Meselson, M. An Unstable Intermediate Carrying Information from Genes to Ribosomes for Protein Synthesis. Nature 1961, 190, 576–581.
- Jacob, F.; Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961, 3, 318–356.
- Scherrer, K.; Darnell, J.E. Sedimentation characteristics of rapidly labelled RNA from HeLa cells. Biochem. Biophys. Res. Commun. 1962, 7, 486–490.
- Scherrer, K.; Latham, H.; Darnell, J.E. Demonstration of an unstable RNA and of a precursor to ribosomal RNA in HeLa cells. Proc. Natl. Acad. Sci. 1963, 49, 240–248.
- Hoagland, M.B.; Stephenson, M.L.; Scott, J.F.; Hecht, L.I.; Zamecnik, P.C. A Soluble Ribonucleic Acid Intermediate in Protein Synthesis. J. Biol. Chem. 1958, 231, 241–257.
- Weinberg, R.A.; Penman, S. Small molecular weight monodisperse nuclear RNA. J. Mol. Biol. 1968, 38, 289–304.
- Wassarman, D.; Steitz, J. Interactions of small nuclear RNA’s with precursor messenger RNA during in vitro splicing. Science 1992, 257, 1918–1925.
- Reddy, R.; Busch, H. Small Nuclear RNAs: RNA Sequences, Structure, and Modifications. In Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles; Springer: Berlin/Heidelberg, Germany, 1988; pp. 1–37.
- Bachellerie, J.P.; Cavaillé, J.; Hüttenhofer, A. The expanding snoRNA world. Biochimie 2002, 84, 775–790.
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854.
- Cox, D.N.; Chao, A.; Baker, J.; Chang, L.; Qiao, D.; Lin, H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998, 12, 3715–3727.
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811.
- Hamilton, A.J.; Baulcombe, D.C. A Species of Small Antisense RNA in Posttranscriptional Gene Silencing in Plants. Science 1999, 286, 950–952.
- Li, M.; Li, J.; Ding, X.; He, M.; Cheng, S.-Y. microRNA and Cancer. AAPS J. 2010, 12, 309–317.
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402.
- Fatica, A.; Rosa, A.; Ballarino, M.; Marchis, M.L.D.; Rasmussen, K.D.; Bozzoni, I. Role of microRNAs in myeloid differentiation. Biochem. Soc. Trans. 2008, 36, 1201–1205.
- Deiuliis, J.A. MicroRNAs as regulators of metabolic disease: Pathophysiologic significance and emerging role as biomarkers and therapeutics. Int. J. Obes. 2016, 40, 88–101.
- Abdul-Muneer, P.M. MicroRNA in the Pathophysiology of CNS Injury: Implication in Neuroregenerative Medicine. CNS Neurosci. Ther. 2016, 22, 543–545.
- De Benedittis, G.; Ciccacci, C.; Latini, A.; Novelli, L.; Novelli, G.; Borgiani, P. Emerging Role of microRNAs and Long Non-Coding RNAs in Sjögren’s Syndrome. Genes 2021, 12, 903.
- He, X.; Kuang, G.; Wu, Y.; Ou, C. Emerging roles of exosomal miRNAs in diabetes mellitus. Clin. Transl. Med. 2021, 11, e468.
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The Transcriptional Landscape of the Mammalian Genome. Science 2005, 309, 1559–1563.
- Mattick, J.S. The central role of RNA in human development and cognition. FEBS Lett. 2011, 585, 1600–1616.
- Nagano, T.; Fraser, P. No-Nonsense Functions for Long Noncoding RNAs. Cell 2011, 145, 178–181.
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S.; et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012, 22, 1760–1774.
- Abugessaisa, I.; Noguchi, S.; Hasegawa, A.; Harshbarger, J.; Kondo, A.; Lizio, M.; Severin, J.; Carninci, P.; Kawaji, H.; Kasukawa, T. FANTOM5 CAGE profiles of human and mouse reprocessed for GRCh38 and GRCm38 genome assemblies. Sci. Data 2017, 4, 170107.
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res. 2021, 49, D884–D891.
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2013, 15, 7–21.
- Yao, R.-W.; Wang, Y.; Chen, L.-L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551.
- Rinn, J.L.; Chang, H.Y. Long Noncoding RNAs: Molecular Modalities to Organismal Functions. Annu. Rev. Biochem. 2020, 89, 283–308.
- Chen, L.-L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016, 17, 205–211.
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691.
- Braicu, C.; Zimta, A.-A.; Gulei, D.; Olariu, A.; Berindan-Neagoe, I. Comprehensive analysis of circular RNAs in pathological states: Biogenesis, cellular regulation, and therapeutic relevance. Cell. Mol. Life Sci. 2019, 76, 1559–1577.
- Patop, I.L.; Wüst, S.; Kadener, S. Past, present, and future of circRNAs. EMBO J. 2019, 38, e100836.
- Xie, R.; Zhang, Y.; Zhang, J.; Li, J.; Zhou, X. The Role of Circular RNAs in Immune-Related Diseases. Front. Immunol. 2020, 11, 545.
- Goldgraben, M.A.; Russell, R.; Rueda, O.M.; Caldas, C.; Git, A. Double-stranded microRNA mimics can induce length- and passenger strand–dependent effects in a cell type–specific manner. RNA 2016, 22, 193–203.
- Tang, L.; Chen, H.Y.; Hao, N.B.; Tang, B.; Guo, H.; Yong, X.; Dong, H.; Yang, S.M. microRNA inhibitors: Natural and artificial sequestration of microRNA. Cancer Lett. 2017, 407, 139–147.
- Seto, A.G.; Beatty, X.; Lynch, J.M.; Hermreck, M.; Tetzlaff, M.; Duvic, M.; Jackson, A.L. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br. J. Haematol. 2018, 183, 428–444.
- Weinstock, B.A.; Feldman, D.L.; Fornoni, A.; Gross, O.; Kashtan, C.E.; Lagas, S.; Lennon, R.; Miner, J.H.; Rheault, M.N.; Simon, J.F.; et al. Clinical trial recommendations for potential Alport syndrome therapies. Kidney Int. 2020, 97, 1109–1116.
- van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L.; et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017, 18, 1386–1396.
- Huang, C.-K.; Kafert-Kasting, S.; Thum, T. Preclinical and Clinical Development of Noncoding RNA Therapeutics for Cardiovascular Disease. Circ. Res. 2020, 126, 663–678.
- Tsimikas, S.; Karwatowska-Prokopczuk, E.; Gouni-Berthold, I.; Tardif, J.-C.; Baum, S.J.; Steinhagen-Thiessen, E.; Shapiro, M.D.; Stroes, E.S.; Moriarty, P.M.; Nordestgaard, B.G.; et al. Lipoprotein(a) Reduction in Persons with Cardiovascular Disease. N. Engl. J. Med. 2020, 382, 244–255.
- Gallant-Behm, C.L.; Piper, J.; Dickinson, B.A.; Dalby, C.M.; Pestano, L.A.; Jackson, A.L. A synthetic microRNA-92a inhibitor (MRG-110) accelerates angiogenesis and wound healing in diabetic and nondiabetic wounds. Wound Repair Regen. 2018, 26, 311–323.
- Cirak, S.; Arechavala-Gomeza, V.; Guglieri, M.; Feng, L.; Torelli, S.; Anthony, K.; Abbs, S.; Garralda, M.E.; Bourke, J.; Wells, D.J.; et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: An open-label, phase 2, dose-escalation study. Lancet 2011, 378, 595–605.
- Voit, T.; Topaloglu, H.; Straub, V.; Muntoni, F.; Deconinck, N.; Campion, G.; De Kimpe, S.J.; Eagle, M.; Guglieri, M.; Hood, S.; et al. Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): An exploratory, randomised, placebo-controlled phase 2 study. Lancet Neurol. 2014, 13, 987–996.
- Clemens, P.R.; Rao, V.K.; Connolly, A.M.; Harper, A.D.; Mah, J.K.; Smith, E.C.; McDonald, C.M.; Zaidman, C.M.; Morgenroth, L.P.; Osaki, H.; et al. Safety, Tolerability, and Efficacy of Viltolarsen in Boys With Duchenne Muscular Dystrophy Amenable to Exon 53 Skipping: A Phase 2 Randomized Clinical Trial. JAMA Neurol. 2020, 77, 982–991.
- Frank, D.E.; Schnell, F.J.; Akana, C.; El-Husayni, S.H.; Desjardins, C.A.; Morgan, J.; Charleston, J.S.; Sardone, V.; Domingos, J.; Dickson, G.; et al. Increased dystrophin production with golodirsen in patients with Duchenne muscular dystrophy. Neurology 2020, 94, e2270–e2282.
- Shirley, M. Casimersen: First Approval. Drugs 2021, 81, 875–879.
- Gan, L.-M.; Lagerström-Fermér, M.; Carlsson, L.G.; Arfvidsson, C.; Egnell, A.-C.; Rudvik, A.; Kjaer, M.; Collén, A.; Thompson, J.D.; Joyal, J.; et al. Intradermal delivery of modified mRNA encoding VEGF-A in patients with type 2 diabetes. Nat. Commun. 2019, 10, 871.
- Kaur, K.; Zangi, L. Modified mRNA as a Therapeutic Tool for the Heart. Cardiovasc. Drugs Ther. 2020, 34, 871–880.
- Eulalio, A.; Mano, M.; Ferro, M.D.; Zentilin, L.; Sinagra, G.; Zacchigna, S.; Giacca, M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 2012, 492, 376–381.
- Gabisonia, K.; Prosdocimo, G.; Aquaro, G.D.; Carlucci, L.; Zentilin, L.; Secco, I.; Ali, H.; Braga, L.; Gorgodze, N.; Bernini, F.; et al. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature 2019, 569, 418–422.
- He, S.; Wang, L.; Miao, L.; Wang, T.; Du, F.; Zhao, L.; Wang, X. Receptor Interacting Protein Kinase-3 Determines Cellular Necrotic Response to TNF-α. Cell 2009, 137, 1100–1111.
- Zhang, D.-Y.; Wang, B.-J.; Ma, M.; Yu, K.; Zhang, Q.; Zhang, X.-W. MicroRNA-325-3p protects the heart after myocardial infarction by inhibiting RIPK3 and programmed necrosis in mice. BMC Mol. Biol. 2019, 20, 17.
- Seale, P.; Sabourin, L.A.; Girgis-Gabardo, A.; Mansouri, A.; Gruss, P.; Rudnicki, M.A. Pax7 Is Required for the Specification of Myogenic Satellite Cells. Cell 2000, 102, 777–786.
- von Maltzahn, J.; Jones, A.E.; Parks, R.J.; Rudnicki, M.A. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc. Natl. Acad. Sci. USA 2013, 110, 16474–16479.
- Chen, J.-F.; Tao, Y.; Li, J.; Deng, Z.; Yan, Z.; Xiao, X.; Wang, D.-Z. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J. Cell Biol. 2010, 190, 867–879.
- Fortier, M.; Figeac, N.; White, R.B.; Knopp, P.; Zammit, P.S. Sphingosine-1-phosphate receptor 3 influences cell cycle progression in muscle satellite cells. Dev. Biol. 2013, 382, 504–516.
- Zhai, L.; Wu, R.; Han, W.; Zhang, Y.; Zhu, D. miR-127 enhances myogenic cell differentiation by targeting S1PR3. Cell Death Dis. 2017, 8, e2707.
- Barclay, R.D.; Burd, N.A.; Tyler, C.; Tillin, N.A.; Mackenzie, R.W. The Role of the IGF-1 Signaling Cascade in Muscle Protein Synthesis and Anabolic Resistance in Aging Skeletal Muscle. Front. Nutr. 2019, 6, 146.
- Schiaffino, S.; Mammucari, C. Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: Insights from genetic models. Skelet. Muscle 2011, 1, 4.
- Li, J.; Chan, M.C.; Yu, Y.; Bei, Y.; Chen, P.; Zhou, Q.; Cheng, L.; Chen, L.; Ziegler, O.; Rowe, G.C.; et al. miR-29b contributes to multiple types of muscle atrophy. Nat. Commun. 2017, 8, 15201.
- Stephenson, M.L.; Zamecnik, P.C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc. Natl. Acad. Sci. USA 1978, 75, 285–288.
- Wu, H.; Lima, W.F.; Zhang, H.; Fan, A.; Sun, H.; Crooke, S.T. Determination of the Role of the Human RNase H1 in the Pharmacology of DNA-like Antisense Drugs. J. Biol. Chem. 2004, 279, 17181–17189.
- Dulla, K.; Aguila, M.; Lane, A.; Jovanovic, K.; Parfitt, D.A.; Schulkens, I.; Chan, H.L.; Schmidt, I.; Beumer, W.; Vorthoren, L.; et al. Splice-Modulating Oligonucleotide QR-110 Restores CEP290 mRNA and Function in Human c.2991+1655A>G LCA10 Models. Mol. Ther.-Nucleic Acids 2018, 12, 730–740.
- Baker, B.F.; Lot, S.S.; Condon, T.P.; Cheng-Flournoy, S.; Lesnik, E.A.; Sasmor, H.M.; Bennett, C.F. 2′-O-(2-Methoxy)ethyl-modified Anti-intercellular Adhesion Molecule 1 (ICAM-1) Oligonucleotides Selectively Increase the ICAM-1 mRNA Level and Inhibit Formation of the ICAM-1 Translation Initiation Complex in Human Umbilical Vein Endothelial Cells. J. Biol. Chem. 1997, 272, 11994–12000.
- Liang, X.; Shen, W.; Sun, H.; Migawa, M.T.; Vickers, T.A.; Crooke, S.T. Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat. Biotechnol. 2016, 34, 875–880.
- Glascock, J.; Lenz, M.; Hobby, K.; Jarecki, J. Cure SMA and our patient community celebrate the first approved drug for SMA. Gene Ther. 2017, 24, 498–500.
- Hua, Y.; Vickers, T.A.; Baker, B.F.; Bennett, C.F.; Krainer, A.R. Enhancement of SMN2 Exon 7 Inclusion by Antisense Oligonucleotides Targeting the Exon. PLOS Biol. 2007, 5, e73.
- Lim, K.R.Q.; Maruyama, R.; Yokota, T. Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Des. Devel. Ther. 2017, 11, 533–545.
- Himič, V.; Davies, K.E. Evaluating the potential of novel genetic approaches for the treatment of Duchenne muscular dystrophy. Eur. J. Hum. Genet. 2021, 29, 1369–1376.
- Bladen, C.L.; Salgado, D.; Monges, S.; Foncuberta, M.E.; Kekou, K.; Kosma, K.; Dawkins, H.; Lamont, L.; Roy, A.J.; Chamova, T.; et al. The TREAT-NMD DMD Global Database: Analysis of More than 7,000 Duchenne Muscular Dystrophy Mutations. Hum. Mutat. 2015, 36, 395–402.
- Tasfaout, H.; Cowling, B.S.; Laporte, J. Centronuclear myopathies under attack: A plethora of therapeutic targets. J. Neuromuscul. Dis. 2018, 5, 387–406.
- Buono, S.; Ross, J.A.; Tasfaout, H.; Levy, Y.; Kretz, C.; Tayefeh, L.; Matson, J.; Guo, S.; Kessler, P.; Monia, B.P.; et al. Reducing dynamin 2 (DNM2) rescues DNM2-related dominant centronuclear myopathy. Proc. Natl. Acad. Sci. USA 2018, 115, 11066–11071.
- Muñoz, X.M.; Kretz, C.; Silva-Rojas, R.; Ochala, J.; Menuet, A.; Romero, N.B.; Cowling, B.S.; Laporte, J. Physiological impact and disease reversion for the severe form of centronuclear myopathy linked to dynamin. JCI Insight 2020, 5, e137899.
- Day, J.W.; Ranum, L.P.W. RNA pathogenesis of the myotonic dystrophies. Neuromuscul. Disord. 2005, 15, 5–16.
- Mahadevan, M.; Tsilfidis, C.; Sabourin, L.; Shutler, G.; Amemiya, C.; Jansen, G.; Neville, C.; Narang, M.; Barceló, J.; O’Hoy, K.; et al. Myotonic dystrophy mutation: An unstable CTG repeat in the 3′ untranslated region of the gene. Science 1992, 255, 1253–1255.
- Taneja, K.L.; McCurrach, M.; Schalling, M.; Housman, D.; Singer, R.H. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 1995, 128, 995–1002.
- Echeverria, G.V.; Cooper, T.A. RNA-binding proteins in microsatellite expansion disorders: Mediators of RNA toxicity. Brain Res. 2012, 1462, 100.
- Jauvin, D.; Chrétien, J.; Pandey, S.K.; Martineau, L.; Revillod, L.; Bassez, G.; Lachon, A.; McLeod, A.R.; Gourdon, G.; Wheeler, T.M.; et al. Targeting DMPK with Antisense Oligonucleotide Improves Muscle Strength in Myotonic Dystrophy Type 1 Mice. Mol. Ther. Nucleic Acids 2017, 7, 465.
- Yadava, R.S.; Yu, Q.; Mandal, M.; Rigo, F.; Bennett, C.F.; Mahadevan, M.S. Systemic therapy in an RNA toxicity mouse model with an antisense oligonucleotide therapy targeting a non-CUG sequence within the DMPK 3′UTR RNA. Hum. Mol. Genet. 2020, 29, 1440–1453.
- Yadava, R.S.; Mandal, M.; Giese, J.M.; Rigo, F.; Bennett, C.F.; Mahadevan, M.S. Modeling muscle regeneration in RNA toxicity mice. Hum. Mol. Genet. 2021, 30, 1111–1130.
- Pratt, A.J.; MacRae, I.J. The RNA-induced Silencing Complex: A Versatile Gene-silencing Machine. J. Biol. Chem. 2009, 284, 17897–17901.
- Solomon, S.D.; McMurray, J.J.V.; Anand, I.S.; Ge, J.; Lam, C.S.P.; Maggioni, A.P.; Martinez, F.; Packer, M.; Pfeffer, M.A.; Pieske, B.; et al. Angiotensin–Neprilysin Inhibition in Heart Failure with Preserved Ejection Fraction. N. Engl. J. Med. 2019, 381, 1609–1620.
- Wooddell, C.I.; Gehring, A.J.; Yuen, M.-F.; Given, B.D. RNA Interference Therapy for Chronic Hepatitis B Predicts the Importance of Addressing Viral Integration When Developing Novel Cure Strategies. Viruses 2021, 13, 581.
- Golan, T.; Khvalevsky, E.Z.; Hubert, A.; Gabai, R.M.; Hen, N.; Segal, A.; Domb, A.; Harari, G.; David, E.B.; Raskin, S.; et al. RNAi therapy targeting KRAS in combination with chemotherapy for locally advanced pancreatic cancer patients. Oncotarget 2015, 6, 24560–24570.
- El Dika, I.; Lim, H.Y.; Yong, W.P.; Lin, C.-C.; Yoon, J.-H.; Modiano, M.; Freilich, B.; Choi, H.J.; Chao, T.-Y.; Kelley, R.K.; et al. An Open-Label, Multicenter, Phase I, Dose Escalation Study with Phase II Expansion Cohort to Determine the Safety, Pharmacokinetics, and Preliminary Antitumor Activity of Intravenous TKM-080301 in Subjects with Advanced Hepatocellular Carcinoma. Oncologist 2019, 24, 747-e218.
- Stiekema, L.C.A.; Prange, K.H.M.; Hoogeveen, R.M.; Verweij, S.L.; Kroon, J.; Schnitzler, J.G.; Dzobo, K.E.; Cupido, A.J.; Tsimikas, S.; Stroes, E.S.G.; et al. Potent lipoprotein(a) lowering following apolipoprotein(a) antisense treatment reduces the pro-inflammatory activation of circulating monocytes in patients with elevated lipoprotein(a). Eur. Heart J. 2020, 41, 2262–2271.
- Dammes, N.; Peer, D. Paving the Road for RNA Therapeutics. Trends Pharmacol. Sci. 2020, 41, 755–775.
- Apte, R.S.; Chen, D.S.; Ferrara, N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019, 176, 1248–1264.
- Zangi, L.; Lui, K.O.; von Gise, A.; Ma, Q.; Ebina, W.; Ptaszek, L.M.; Später, D.; Xu, H.; Tabebordbar, M.; Gorbatov, R.; et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013, 31, 898–907.
- Lui, K.O.; Zangi, L.; Silva, E.A.; Bu, L.; Sahara, M.; Li, R.A.; Mooney, D.J.; Chien, K.R. Driving vascular endothelial cell fate of human multipotent Isl1 + heart progenitors with VEGF modified mRNA. Cell Res. 2013, 23, 1172–1186.
- Carlsson, L.; Clarke, J.C.; Yen, C.; Gregoire, F.; Albery, T.; Billger, M.; Egnell, A.-C.; Gan, L.-M.; Jennbacken, K.; Johansson, E.; et al. Biocompatible, Purified VEGF-A mRNA Improves Cardiac Function after Intracardiac Injection 1 Week Post-myocardial Infarction in Swine. Mol. Ther.-Methods Clin. Dev. 2018, 9, 330–346.
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358.
- Song, C.; Zhang, J.; Qi, H.; Feng, C.; Chen, Y.; Cao, Y.; Ba, L.; Ai, B.; Wang, Q.; Huang, W.; et al. The global view of mRNA-related ceRNA cross-talks across cardiovascular diseases. Sci. Rep. 2017, 7, 10185.
- Ala, U. Competing Endogenous RNAs, Non-Coding RNAs and Diseases: An Intertwined Story. Cells 2020, 9, 1574.
- Ballarino, M.; Morlando, M.; Fatica, A.; Bozzoni, I. Non-coding RNAs in muscle differentiation and musculoskeletal disease. J. Clin. Invest. 2016, 126, 2021–2030.
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA. Cell 2011, 147, 358–369.
- Shen, H.; McElhinny, A.S.; Cao, Y.; Gao, P.; Liu, J.; Bronson, R.; Griffin, J.D.; Wu, L. The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 2006, 20, 675.
- Cai, B.; Ma, W.; Ding, F.; Zhang, L.; Huang, Q.; Wang, X.; Hua, B.; Xu, J.; Li, J.; Bi, C.; et al. The Long Noncoding RNA CAREL Controls Cardiac Regeneration. J. Am. Coll. Cardiol. 2018, 72, 534–550.
- Qu, X.; Du, Y.; Shu, Y.; Gao, M.; Sun, F.; Luo, S.; Yang, T.; Zhan, L.; Yuan, Y.; Chu, W.; et al. MIAT Is a Pro-fibrotic Long Non-coding RNA Governing Cardiac Fibrosis in Post-infarct Myocardium. Sci. Rep. 2017, 7, 42657.
- Zhang, Z.-K.; Li, J.; Guan, D.; Liang, C.; Zhuo, Z.; Liu, J.; Lu, A.; Zhang, G.; Zhang, B.-T. Long Noncoding RNA lncMUMA Reverses Established Skeletal Muscle Atrophy following Mechanical Unloading. Mol. Ther. 2018, 26, 2669–2680.
- Zhu, M.; Liu, J.; Xiao, J.; Yang, L.; Cai, M.; Shen, H.; Chen, X.; Ma, Y.; Hu, S.; Wang, Z.; et al. Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat. Commun. 2017, 8, 14718.
- Si, X.; Zheng, H.; Wei, G.; Li, M.; Li, W.; Wang, H.; Guo, H.; Sun, J.; Li, C.; Zhong, S.; et al. circRNA Hipk3 Induces Cardiac Regeneration after Myocardial Infarction in Mice by Binding to Notch1 and miR-133a. Mol. Ther.-Nucleic Acids 2020, 21, 636–655.
- Huang, S.; Li, X.; Zheng, H.; Si, X.; Li, B.; Wei, G.; Li, C.; Chen, Y.; Chen, Y.; Liao, W.; et al. Loss of Super-Enhancer-Regulated circRNA Nfix Induces Cardiac Regeneration After Myocardial Infarction in Adult Mice. Circulation 2019, 139, 2857–2876.
- Wang, K.; Long, B.; Liu, F.; Wang, J.-X.; Liu, C.-Y.; Zhao, B.; Zhou, L.-Y.; Sun, T.; Wang, M.; Yu, T.; et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016, 37, 2602–2611.
- Lavenniah, A.; Luu, T.D.A.; Li, Y.P.; Lim, T.B.; Jiang, J.; Ackers-Johnson, M.; Foo, R.S.-Y. Engineered Circular RNA Sponges Act as miRNA Inhibitors to Attenuate Pressure Overload-Induced Cardiac Hypertrophy. Mol. Ther. 2020, 28, 1506–1517.
- Li, J.; Yang, T.; Tang, H.; Sha, Z.; Chen, R.; Chen, L.; Yu, Y.; Rowe, G.C.; Das, S.; Xiao, J. Inhibition of lncRNA MAAT Controls Multiple Types of Muscle Atrophy by cis- and trans-Regulatory Actions. Mol. Ther. 2021, 29, 1102–1119.
- Zhou, L.; Sun, K.; Zhao, Y.; Zhang, S.; Wang, X.; Li, Y.; Lu, L.; Chen, X.; Chen, F.; Bao, X.; et al. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat. Commun. 2015, 6, 10026.
- Ponnusamy, M.; Liu, F.; Zhang, Y.-H.; Li, R.-B.; Zhai, M.; Liu, F.; Zhou, L.-Y.; Liu, C.-Y.; Yan, K.-W.; Dong, Y.-H.; et al. Long Noncoding RNA CPR (Cardiomyocyte Proliferation Regulator) Regulates Cardiomyocyte Proliferation and Cardiac Repair. Circulation 2019, 139, 2668–2684.
- Cipriano, A.; Macino, M.; Buonaiuto, G.; Santini, T.; Biferali, B.; Peruzzi, G.; Colantoni, A.; Mozzetta, C.; Ballarino, M. Epigenetic regulation of wnt7b expression by the cis-acting long noncoding rna lnc-rewind in muscle stem cells. Elife 2021, 10, e54782.
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307.
- Alvarez, S.; Díaz, M.; Flach, J.; Rodriguez-Acebes, S.; López-Contreras, A.J.; Martínez, D.; Cañamero, M.; Fernández-Capetillo, O.; Isern, J.; Passegué, E.; et al. Replication stress caused by low MCM expression limits fetal erythropoiesis and hematopoietic stem cell functionality. Nat. Commun. 2015, 6, 8548.
- Wang, H.; Hertlein, E.; Bakkar, N.; Sun, H.; Acharyya, S.; Wang, J.; Carathers, M.; Davuluri, R.; Guttridge, D.C. NF-κB Regulation of YY1 Inhibits Skeletal Myogenesis through Transcriptional Silencing of Myofibrillar Genes. Mol. Cell. Biol. 2007, 27, 4374–4387.
- Ballarino, M.; Cazzella, V.; D’Andrea, D.; Grassi, L.; Bisceglie, L.; Cipriano, A.; Santini, T.; Pinnarò, C.; Morlando, M.; Tramontano, A.; et al. Novel Long Noncoding RNAs (lncRNAs) in Myogenesis: A miR-31 Overlapping lncRNA Transcript Controls Myoblast Differentiation. Mol. Cell. Biol. 2015, 35, 728–736.
- Tzimagiorgis, G.; Michailidou, E.Z.; Kritis, A.; Markopoulos, A.K.; Kouidou, S. Recovering circulating extracellular or cell-free RNA from bodily fluids. Cancer Epidemiol. 2011, 35, 580–589.
- El-Mogy, M.; Lam, B.; Haj-Ahmad, T.A.; McGowan, S.; Yu, D.; Nosal, L.; Rghei, N.; Roberts, P.; Haj-Ahmad, Y. Diversity and signature of small RNA in different bodily fluids using next generation sequencing. BMC Genomics 2018, 19, 408.
- Cacchiarelli, D.; Legnini, I.; Martone, J.; Cazzella, V.; D’Amico, A.; Bertini, E.; Bozzoni, I. miRNAs as serum biomarkers for Duchenne muscular dystrophy. EMBO Mol. Med. 2011, 3, 258–265.
- Zaharieva, I.T.; Calissano, M.; Scoto, M.; Preston, M.; Cirak, S.; Feng, L.; Collins, J.; Kole, R.; Guglieri, M.; Straub, V.; et al. Dystromirs as Serum Biomarkers for Monitoring the Disease Severity in Duchenne Muscular Dystrophy. PLoS One 2013, 8, e80263.
- Coenen-Stass, A.M.L.; Betts, C.A.; Lee, Y.F.; Mäger, I.; Turunen, M.P.; EL Andaloussi, S.; Morgan, J.E.; Wood, M.J.A.; Roberts, T.C. Selective release of muscle-specific, extracellular microRNAs during myogenic differentiation. Hum. Mol. Genet. 2016, 25, 3960–3974.
- Coenen-Stass, A.M.L.; Sork, H.; Gatto, S.; Godfrey, C.; Bhomra, A.; Krjutškov, K.; Hart, J.R.; Westholm, J.O.; O’Donovan, L.; Roos, A.; et al. Comprehensive RNA-Sequencing Analysis in Serum and Muscle Reveals Novel Small RNA Signatures with Biomarker Potential for DMD. Mol. Ther.-Nucleic Acids 2018, 13, 1–15.
- Perfetti, A.; Greco, S.; Cardani, R.; Fossati, B.; Cuomo, G.; Valaperta, R.; Ambrogi, F.; Cortese, A.; Botta, A.; Mignarri, A.; et al. Validation of plasma microRNAs as biomarkers for myotonic dystrophy type 1. Sci. Rep. 2016, 6, 38174.




