
Video Upload Options
Shiga toxins (Stxs) are classic bacterial toxins and major virulence factors of toxigenic Shigella dysenteriae and enterohemorrhagic Escherichia coli (EHEC). These toxins recognize a glycosphingolipid globotriaosylceramide (Gb3/CD77) as their receptor and inhibit protein synthesis in cells by cleaving 28S ribosomal RNA. They are the major cause of life-threatening complications such as hemolytic uremic syndrome (HUS), associated with severe cases of EHEC infection, which is the leading cause of acute kidney injury in children.
1. Introduction
Shiga toxin (Stx) was named after Japanese microbiologist Kiyoshi Shiga, who identified and characterized Shigella dysenteriae in 1897 [1]. Among Shigella dysenteriae strains, serotype 1 is the toxigenic one that expresses Stx [2]. In 1977, a cytotoxic toxin from Escherichia coli (E. coli) isolates was discovered by Konowalchuck et al., initially named verotoxin or E. coli cytotoxin for its ability to kill cultured Vero cells [3]. By the early 1980s, O’Brien et al. recognized that E. coli isolates express toxins highly similar to Stx; they called them Shiga-like toxins and designated the E. coli isolates Shiga-like toxin-producing E. coli (STEC) [4]. Among STEC, the strains associated with human diseases are also known as enterohemorrhagic E. coli (EHEC). It was soon realized that these toxins belong to the same Stx family and can be divided into two serotypes: Stx1 is almost identical to the prototype Stx in Shigella dysenteriae, while Stx2 shares ~56% protein sequence identity with Stx [5].
Each year, in the United States, there are approximately 265,000 cases of EHEC infection [6], which usually starts with diarrhea and can develop into dysentery and hemorrhagic colitis. In severe cases, life-threatening complications such as hemolytic uremic syndrome (HUS) and neurological disorders can occur. HUS is most commonly associated with EHEC serotype O157:H7 infections, and Stx is the major cause [7]. HUS has the highest incidence in children and the elderly [8][9] and is the major reason for acute kidney injury in children [10]. Patients have typically developed irreversible vascular damage by the time symptoms appear, and there is no specific treatment [7].
Investigations over the past 40 years have elucidated the molecular mechanism underlying the toxicity of Stx, established its central role in pathogenesis, and shed light on many key cellular functions. Furthermore, Stx and Stx fragments have also been utilized as tools for biomedical applications. Many topics in Stx biology and applications have been covered by excellent in-depth reviews in recent years [9][11][12][13][14][15][16][17][18][19][20][21][22].
2. Stx Structure and Function
Stx is an AB5 toxin, comprising an enzymatic A subunit (Stx-A, 32 kDa) and five identical B subunits (Stx-B, 7.7 kDa) that form a pentamer. The A subunit connects to the pentamer noncovalently by inserting its C-terminal region into the central hole [23][24] (Figure 1). The A subunit is an RNA N-glycosidase, which inhibits protein synthesis by cleaving 28S ribosomal RNA [25][26][27]. The B subunit is responsible for binding to receptors. The glycosphingolipid globotriaosylceramide (Gb3, also known as CD77) is the major receptor of Stx [28]. Crystal structural studies using Stx1-B and a trisaccharide analog of Gb3 revealed three Gb3 binding sites per B subunit (Figure 1). Thus, one Stx can bind up to 15 Gb3 simultaneously [29]. After clustering Gb3 on cell surfaces, Stx is internalized by clathrin-dependent and -independent endocytosis pathways [30][31], retrogradely sorted into the trans-Golgi network (TGN) and further into the endoplasmic reticulum (ER), bypassing the late endocytic pathway [32][33][34][35][36]. The A subunit is further processed by host furin and furin-like proteases (cleaving between R251 and M252 of Stx1 and between R250 and A251 of Stx2) into the enzymatic piece A1 (27.5 kDa) and the linker piece A2 (4.5 kDa), connecting to the B subunit. The A1 and A2 domains remain connected by a single disulfide bond, which is reduced within the ER. The A1 domain is then released from the ER into the cytosol through the ER-associated protein degradation (ERAD) pathway to inhibit protein synthesis [14][37][38].
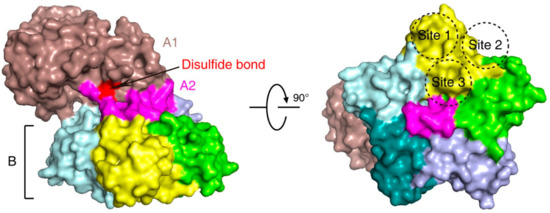
Figure 1. The structure of Shiga toxin: an A subunit, which is cleaved into an enzymatic piece A1 (colored brown) and a linker piece A2 (colored magenta), and five B subunits (PDB: 4M1U). The disulfide bond connecting A1 and A2 pieces is colored red. In the bottom view (right panel), three receptor binding sites on one B subunit (colored yellow) are shown.
3. Stx Subtypes
Stx1 and Stx2 each have multiple subtypes classified based on protein sequence variations. Three subtypes of Stx1 (Stx1a, Stx1c, and Stx1d) and eleven subtypes of Stx2 (Stx2a–Stx2k) have been reported so far [39][40][41][42]. Some STEC strains express only one type of Stx, while others may express multiple types simultaneously [39]. Protein stability, toxin potency, receptor preference, symptom severity, and related host reservoirs may vary among subtypes [39][43]. For example, strains that produce Stx2a, Stx2c, and Stx2d are often associated with colitis and HUS in human infections. STEC producing Stx2b, Stx2e, Stx2f, and Stx2g are usually associated with animal infections, such as in deer, pigs, pigeons, and cattle [17][41][44][45]. Stx2e is the common subtype causing the edema disease of swine [46]. Strains producing Stx2f are frequently found in pigeons and other birds [47][48][49]. In 2018, Stx2h and Stx2i were identified in STEC strains isolated from wild marmots and shrimp, respectively [40][50]. Two additional new subtypes, Stx2j and Stx2k, have recently been reported but have yet to be broadly accepted. Stx2k was isolated from bacterial strains of various sources, including animals (goats and pigs), raw meat (beef and mutton), and human patients with diarrhea [41][42].
Stx1 and most Stx2 subtypes recognize the Gb3 receptor. However, Stx2e binds preferentially to globotetraosylceramide (Gb4), which is synthesized by adding N-acetylgalactosamine to Gb3 [51]. Toxicity differences among Stx subtypes are partially ascribed to differences in receptor binding, and it is believed that the low dissociation rate of Stx2a with receptors may lead to greater toxicity due to longer toxin uptake [52]. This may be one reason why Stx2a, among all Stx2 subtypes, is implicated in most HUS cases [53].
References
- Trofa, A.F.; Ueno-Olsen, H.; Oiwa, R.; Yoshikawa, M. Dr. Kiyoshi Shiga: Discoverer of the dysentery bacillus. Clin. Infect. Dis. 1999, 29, 1303–1306.
- Zaidi, M.B.; Estrada-Garcia, T. Shigella: A Highly Virulent and Elusive Pathogen. Curr. Trop. Med. Rep. 2014, 1, 81–87.
- Konowalchuk, J.; Speirs, J.I.; Stavric, S. Vero response to a cytotoxin of Escherichia coli. Infect. Immun. 1977, 18, 775–779.
- O’Brien, A.D.; Newland, J.W.; Miller, S.F.; Holmes, R.K.; Smith, H.W.; Formal, S.B. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 1984, 226, 694–696.
- Jackson, M.P.; Neill, R.J.; O’Brien, A.D.; Holmes, R.K.; Newland, J.W. Nucleotide sequence analysis and comparison of the structural genes for Shiga-like toxin I and Shiga-like toxin II encoded by bacteriophages from Escherichia coli 933. FEMS Microbiol. Lett. 1987, 44, 109–114.
- Centers for Disease Control and Prevention (CDC). National Shiga Toxin-Producing Escherichia coli (STEC) Surveillance Overview. Atlanta, Georgia: US Department of Health and Human Services, CDC. 2012. Available online: (accessed on 17 March 2021).
- Tarr, P.I.; Gordon, C.A.; Chandler, W.L. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 2005, 365, 1073–1086.
- Boyce, T.G.; Swerdlow, D.L.; Griffin, P.M. Escherichia coli O157:H7 and the hemolytic-uremic syndrome. N. Engl. J. Med. 1995, 333, 364–368.
- Joseph, A.; Cointe, A.; Mariani Kurkdjian, P.; Rafat, C.; Hertig, A. Shiga Toxin-Associated Hemolytic Uremic Syndrome: A Narrative Review. Toxins 2020, 12, 67.
- Williams, D.M.; Sreedhar, S.S.; Mickell, J.J.; Chan, J.C. Acute kidney failure: A pediatric experience over 20 years. Arch. Pediatr. Adolesc. Med. 2002, 156, 893–900.
- Johannes, L.; Romer, W. Shiga toxins--from cell biology to biomedical applications. Nat. Rev. Microbiol. 2010, 8, 105–116.
- Obrig, T.G. Escherichia coli Shiga Toxin Mechanisms of Action in Renal Disease. Toxins 2010, 2, 2769–2794.
- Engedal, N.; Skotland, T.; Torgersen, M.L.; Sandvig, K. Shiga toxin and its use in targeted cancer therapy and imaging. Microb. Biotechnol. 2011, 4, 32–46.
- Bergan, J.; Dyve Lingelem, A.B.; Simm, R.; Skotland, T.; Sandvig, K. Shiga toxins. Toxicon 2012, 60, 1085–1107.
- Obrig, T.G.; Karpman, D. Shiga toxin pathogenesis: Kidney complications and renal failure. Curr. Top. Microbiol. Immunol. 2012, 357, 105–136.
- Pacheco, A.R.; Sperandio, V. Shiga toxin in enterohemorrhagic E.coli: Regulation and novel anti-virulence strategies. Front. Cell Infect. Microbiol. 2012, 2, 81.
- Melton-Celsa, A.R. Shiga Toxin (Stx) Classification, Structure, and Function. Microbiol. Spectr. 2014, 2, 37–53.
- Jeong, Y.J.; Park, S.K.; Yoon, S.J.; Park, Y.J.; Lee, M.S. Experimental In Vivo Models of Bacterial Shiga Toxin-Associated Hemolytic Uremic Syndrome. J. Microbiol. Biotechnol. 2018, 28, 1413–1425.
- Lee, M.S.; Tesh, V.L. Roles of Shiga Toxins in Immunopathology. Toxins 2019, 11, 212.
- Lingwood, C. Verotoxin Receptor-Based Pathology and Therapies. Front. Cell Infect. Microbiol. 2020, 10, 123.
- Luginbuehl, V.; Meier, N.; Kovar, K.; Rohrer, J. Intracellular drug delivery: Potential usefulness of engineered Shiga toxin subunit B for targeted cancer therapy. Biotechnol. Adv. 2018, 36, 613–623.
- Menge, C. Molecular Biology of Escherichia Coli Shiga Toxins’ Effects on Mammalian Cells. Toxins 2020, 12, 345.
- Fraser, M.E.; Chernaia, M.M.; Kozlov, Y.V.; James, M.N. Crystal structure of the holotoxin from Shigella dysenteriae at 2.5 A resolution. Nat. Struct. Biol. 1994, 1, 59–64.
- Stein, P.E.; Boodhoo, A.; Tyrrell, G.J.; Brunton, J.L.; Read, R.J. Crystal structure of the cell-binding B oligomer of verotoxin-1 from E. coli. Nature 1992, 355, 748–750.
- Endo, Y.; Tsurugi, K.; Yutsudo, T.; Takeda, Y.; Ogasawara, T.; Igarashi, K. Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. RNA N-glycosidase activity of the toxins. Eur. J. Biochem. 1988, 171, 45–50.
- Saxena, S.K.; O’Brien, A.D.; Ackerman, E.J. Shiga toxin, Shiga-like toxin II variant, and ricin are all single-site RNA N-glycosidases of 28 S RNA when microinjected into Xenopus oocytes. J. Biol. Chem. 1989, 264, 596–601.
- Ogasawara, T.; Ito, K.; Igarashi, K.; Yutsudo, T.; Nakabayashi, N.; Takeda, Y. Inhibition of protein synthesis by a Vero toxin (VT2 or Shiga-like toxin II) produced by Escherichia coli O157:H7 at the level of elongation factor 1-dependent aminoacyl-tRNA binding to ribosomes. Microb. Pathog. 1988, 4, 127–135.
- Lindberg, A.A.; Brown, J.E.; Stromberg, N.; Westling-Ryd, M.; Schultz, J.E.; Karlsson, K.A. Identification of the carbohydrate receptor for Shiga toxin produced by Shigella dysenteriae type 1. J. Biol. Chem. 1987, 262, 1779–1785.
- Ling, H.; Boodhoo, A.; Hazes, B.; Cummings, M.D.; Armstrong, G.D.; Brunton, J.L.; Read, R.J. Structure of the shiga-like toxin I B-pentamer complexed with an analogue of its receptor Gb3. Biochemistry 1998, 37, 1777–1788.
- Romer, W.; Pontani, L.L.; Sorre, B.; Rentero, C.; Berland, L.; Chambon, V.; Lamaze, C.; Bassereau, P.; Sykes, C.; Gaus, K.; et al. Actin dynamics drive membrane reorganization and scission in clathrin-independent endocytosis. Cell 2010, 140, 540–553.
- Sandvig, K.; van Deurs, B. Endocytosis, intracellular transport, and cytotoxic action of Shiga toxin and ricin. Physiol. Rev. 1996, 76, 949–966.
- Sandvig, K.; Olsnes, S.; Brown, J.E.; Petersen, O.W.; van Deurs, B. Endocytosis from coated pits of Shiga toxin: A glycolipid-binding protein from Shigella dysenteriae 1. J. Cell Biol. 1989, 108, 1331–1343.
- Saint-Pol, A.; Yelamos, B.; Amessou, M.; Mills, I.G.; Dugast, M.; Tenza, D.; Schu, P.; Antony, C.; McMahon, H.T.; Lamaze, C.; et al. Clathrin adaptor epsinR is required for retrograde sorting on early endosomal membranes. Dev. Cell 2004, 6, 525–538.
- Johannes, L.; Popoff, V. Tracing the retrograde route in protein trafficking. Cell 2008, 135, 1175–1187.
- Sandvig, K.; Skotland, T.; van Deurs, B.; Klokk, T.I. Retrograde transport of protein toxins through the Golgi apparatus. Histochem. Cell Biol. 2013, 140, 317–326.
- Mallard, F.; Antony, C.; Tenza, D.; Salamero, J.; Goud, B.; Johannes, L. Direct pathway from early/recycling endosomes to the Golgi apparatus revealed through the study of shiga toxin B-fragment transport. J. Cell Biol. 1998, 143, 973–990.
- Garred, O.; Dubinina, E.; Polesskaya, A.; Olsnes, S.; Kozlov, J.; Sandvig, K. Role of the disulfide bond in Shiga toxin A-chain for toxin entry into cells. J. Biol. Chem. 1997, 272, 11414–11419.
- Fagerquist, C.K.; Sultan, O. Top-down proteomic identification of furin-cleaved alpha-subunit of Shiga toxin 2 from Escherichia coli O157:H7 using MALDI-TOF-TOF-MS/MS. J. Biomed. Biotechnol. 2010, 2010, 123460.
- Scheutz, F.; Teel, L.D.; Beutin, L.; Pierard, D.; Buvens, G.; Karch, H.; Mellmann, A.; Caprioli, A.; Tozzoli, R.; Morabito, S.; et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 2012, 50, 2951–2963.
- Bai, X.; Fu, S.; Zhang, J.; Fan, R.; Xu, Y.; Sun, H.; He, X.; Xu, J.; Xiong, Y. Identification and pathogenomic analysis of an Escherichia coli strain producing a novel Shiga toxin 2 subtype. Sci. Rep. 2018, 8, 6756.
- Hughes, A.C.; Zhang, Y.; Bai, X.; Xiong, Y.; Wang, Y.; Yang, X.; Xu, Q.; He, X. Structural and Functional Characterization of Stx2k, a New Subtype of Shiga Toxin 2. Microorganisms 2019, 8, 4.
- Yang, X.; Bai, X.; Zhang, J.; Sun, H.; Fu, S.; Fan, R.; He, X.; Scheutz, F.; Matussek, A.; Xiong, Y. Escherichia coli strains producing a novel Shiga toxin 2 subtype circulate in China. Int. J. Med. Microbiol. 2020, 310, 151377.
- Fuller, C.A.; Pellino, C.A.; Flagler, M.J.; Strasser, J.E.; Weiss, A.A. Shiga toxin subtypes display dramatic differences in potency. Infect. Immun. 2011, 79, 1329–1337.
- Luna-Gierke, R.E.; Griffin, P.M.; Gould, L.H.; Herman, K.; Bopp, C.A.; Strockbine, N.; Mody, R.K. Outbreaks of non-O157 Shiga toxin-producing Escherichia coli infection: USA. Epidemiol. Infect. 2014, 142, 2270–2280.
- Fraser, M.E.; Fujinaga, M.; Cherney, M.M.; Melton-Celsa, A.R.; Twiddy, E.M.; O’Brien, A.D.; James, M.N. Structure of shiga toxin type 2 (Stx2) from Escherichia coli O157:H7. J. Biol. Chem. 2004, 279, 27511–27517.
- Casanova, N.A.; Redondo, L.M.; Dailoff, G.C.; Arenas, D.; Fernandez Miyakawa, M.E. Overview of the role of Shiga toxins in porcine edema disease pathogenesis. Toxicon 2018, 148, 149–154.
- Farooq, S.; Hussain, I.; Mir, M.A.; Bhat, M.A.; Wani, S.A. Isolation of atypical enteropathogenic Escherichia coli and Shiga toxin 1 and 2f-producing Escherichia coli from avian species in India. Lett. Appl. Microbiol. 2009, 48, 692–697.
- Murakami, K.; Etoh, Y.; Ichihara, S.; Maeda, E.; Takenaka, S.; Horikawa, K.; Narimatsu, H.; Kawano, K.; Kawamura, Y.; Ito, K. Isolation and characteristics of Shiga toxin 2f-producing Escherichia coli among pigeons in Kyushu, Japan. PLoS ONE 2014, 9, e86076.
- Schmidt, H.; Scheef, J.; Morabito, S.; Caprioli, A.; Wieler, L.H.; Karch, H. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 2000, 66, 1205–1208.
- Lacher, D.W.; Gangiredla, J.; Patel, I.; Elkins, C.A.; Feng, P.C. Use of the Escherichia coli Identification Microarray for Characterizing the Health Risks of Shiga Toxin-Producing Escherichia coli Isolated from Foods. J. Food Prot. 2016, 79, 1656–1662.
- Tyrrell, G.J.; Ramotar, K.; Toye, B.; Boyd, B.; Lingwood, C.A.; Brunton, J.L. Alteration of the carbohydrate binding specificity of verotoxins from Gal alpha 1-4Gal to GalNAc beta 1-3Gal alpha 1-4Gal and vice versa by site-directed mutagenesis of the binding subunit. Proc. Natl. Acad. Sci. USA 1992, 89, 524–528.
- Nakajima, H.; Kiyokawa, N.; Katagiri, Y.U.; Taguchi, T.; Suzuki, T.; Sekino, T.; Mimori, K.; Ebata, T.; Saito, M.; Nakao, H.; et al. Kinetic analysis of binding between Shiga toxin and receptor glycolipid Gb3Cer by surface plasmon resonance. J. Biol. Chem. 2001, 276, 42915–42922.
- Friedrich, A.W.; Bielaszewska, M.; Zhang, W.L.; Pulz, M.; Kuczius, T.; Ammon, A.; Karch, H. Escherichia coli harboring Shiga toxin 2 gene variants: Frequency and association with clinical symptoms. J. Infect. Dis. 2002, 185, 74–84.




