
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Marco Vacante | + 270 word(s) | 270 | 2020-09-16 09:44:50 | | | |
| 2 | Dean Liu | Meta information modification | 270 | 2020-09-21 07:43:34 | | | | |
| 3 | Dean Liu | -1 word(s) | 269 | 2020-10-21 08:40:38 | | | | |
| 4 | Dean Liu | Meta information modification | 269 | 2020-10-21 09:04:11 | | | | |
| 5 | Dean Liu | + 1114 word(s) | 1383 | 2020-11-09 09:23:36 | | | | |
| 6 | Dean Liu | Meta information modification | 1383 | 2020-11-09 10:13:45 | | |
Video Upload Options
There is a critical need for new diagnostic and prognostic biomarkers with high specificity and sensitivity in patients with colorectal cancer (CRC). Liquid biopsy could represent the new era for biomarkers detection: the term “liquid biopsy” refers to the isolation of cancer-derived components, such as circulating tumor cells (CTC), circulating tumor DNA (ctDNA), microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and proteins, from peripheral blood or other body fluids (i.e., ascites, urine, pleural effusion, and cerebrospinal fluid), and their genomic or proteomic assessment. Furthermore, exosomes (EXOs) which are membrane-bound extracellular vesicles containing proteins and nucleic acids released in the bloodstream by cancer cells, could represent potential biomarkers.
1. Introduction
Colorectal cancer (CRC) is one of the most common solid cancers in developed countries, with approximately 1.8 million incident cases and 900,000 deaths every year worldwide [1][2]. The burden of CRC is growing in the majority of low- and middle-income countries, probably due to environmental risk factors, such as changes in diet and life-style (i.e., obesity, smoking, alcohol consumption, and suboptimal dietary habits) [3], aging, and urbanization[4][5]. According to the American Cancer Society (ACS), the 5-year survival rate ranges from 90% if CRC is diagnosed at a localized stage to 14% in patients presenting with metastatic disease[6]. Treatment decisions for CRC should take into account the stage of the disease, the general condition, and performance status of the patient, and the molecular characteristics of the tumor[7][8]. The diagnosis of CRC is frequently made using colonoscopy, and confirmed by histological examination of the tumor tissue biopsy. The TNM staging of CRC is based on the depth of invasion of the primary tumor, regional lymph node involvement, and distant metastases, which may contribute to the choice of the most appropriate therapeutic approach, including adjuvant chemotherapy[9]. Surgical resection with lymph node dissection represents the base of curative treatment for localized colon cancer. Patients with stage III colon cancer are treated with adjuvant therapy using the FOLFOX (leucovorin, 5-fluorouracil and oxaliplatin) regimen; however more data are needed to confirm the efficacy of such treatment for rectal cancer patients. Combination of doublet or triplet chemotherapy (i.e., 5-fluorouracil/leucovorin, capecitabine, oxaliplatin, irinotecan) and a targeted agent (i.e., cetuximab, bevacizumab, panitumumab) are routinely used for the treatment of metastatic CRC[10][11]. Histopathological tumor tissue analysis cannot be considered to be a reliable source of clinically helpful prognostic or predictive information for CRC at the individual patient’s level; thus, research is constantly moving towards the identification of more accurate and personalized biomarkers[12]. Indeed, there is a critical need for new diagnostic and prognostic biomarkers with high specificity and sensitivity in patients with CRC[13][14]. In this context, liquid biopsy could represent the new era for biomarkers detection: the term “liquid biopsy” refers to the isolation of cancer-derived components, such as circulating tumor cells (CTC), circulating tumor DNA (ctDNA), microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and proteins, from peripheral blood or other body fluids (i.e., ascites, urine, pleural effusion, and cerebrospinal fluid), and their genomic or proteomic assessment[15][16]. Furthermore, exosomes (EXOs) which are membrane-bound extracellular vesicles containing proteins and nucleic acids released in the bloodstream by cancer cells, could represent potential biomarkers [17][18].
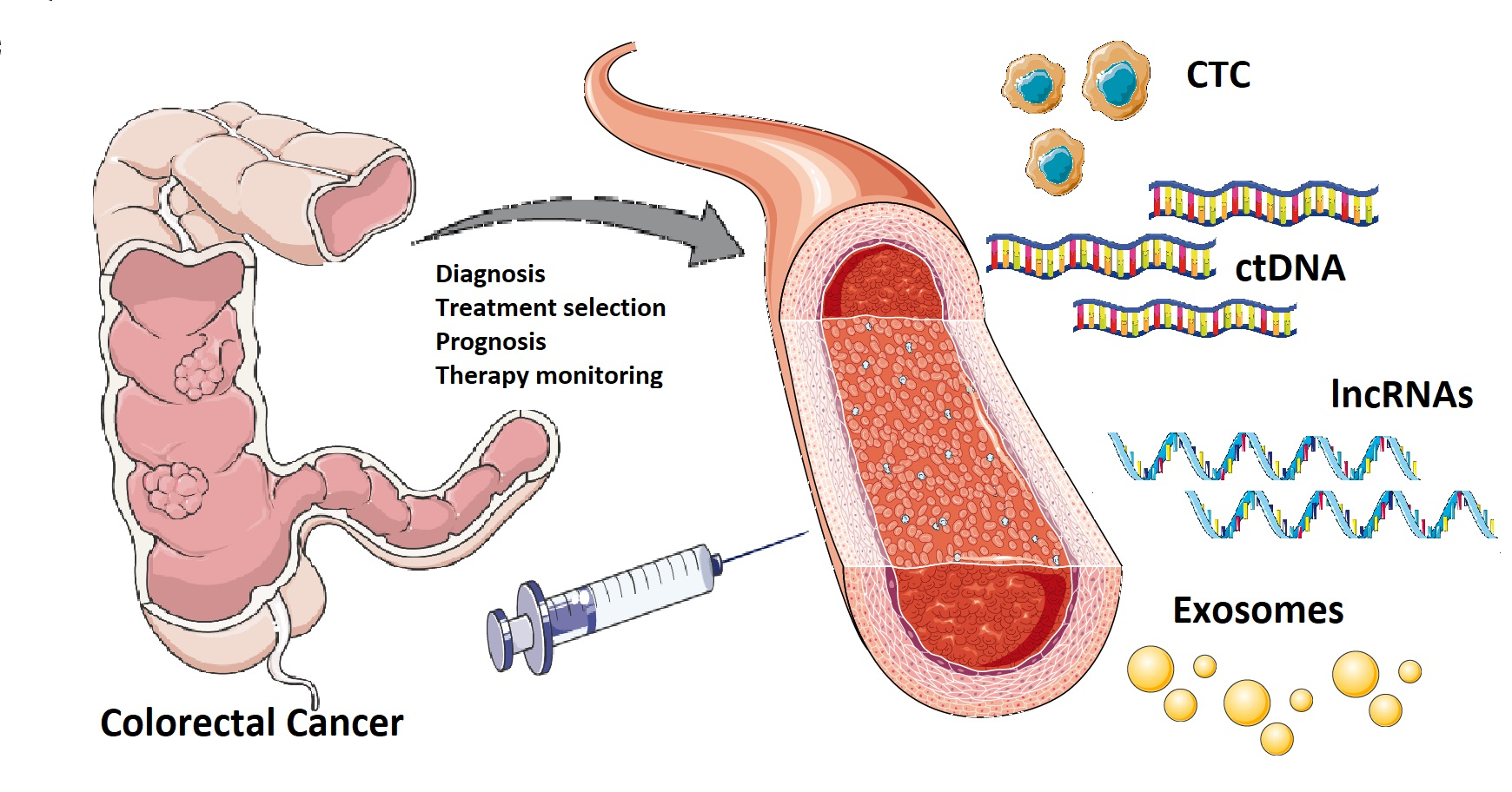
Figure 1. Currently, there is a crucial need for novel diagnostic and prognostic biomarkers with high specificity and sensitivity in patients with colorectal cancer. A “liquid biopsy” is characterized by the isolation of cancer-derived components, such as circulating tumor cells, circulating tumor DNA, microRNAs, long non-coding RNAs, and proteins, from peripheral blood or other body fluids and their genomic or proteomic assessment. The liquid biopsy is a minimally invasive and repeatable technique that could play a significant role in screening and diagnosis, and predict relapse and metastasis, as well as monitoring minimal residual disease and chemotherapy resistance in colorectal cancer patients. However, there are still some practical issues that need to be addressed before liquid biopsy can be widely used in clinical practice. Potential challenges may include low amounts of circulating tumor cells and circulating tumor DNA in samples, lack of pre-analytical and analytical consensus, clinical validation, and regulatory endorsement.
2. Current Issues and Limitations of Liquid Biopsy
Despite all the potential advantages of liquid biopsy in the management of CRC, there are still some practical issues that need to be addressed before it can be widely used in clinical practice[19]. Potential challenges may include low amounts of CTCs and ctDNA in samples, lack of pre-analytical and analytical consensus, clinical validation, regulatory endorsement and cost effectiveness[20][21]. Currently, the use of CTCs in routine diagnostics is limited, mainly due to methodological constraints, such as the lack of an established assessment practice, beyond enumeration[22][23]. The epithelial cell adhesion molecule (EpCAM)-dependent technique was approved by the U.S. Food and Drug Administration (FDA) in 2004, and represents the “gold standard” for CTC isolation in different cancers, including CRC[24]. However, only CTCs that maintain epithelial features can be detected by EpCAM, excluding CTCs with mesenchymal characteristics[25]. On the other hand, ctDNA analysis has been better optimized for routine diagnostic use[26]. The concentration of ctDNA in the peripheral blood depends on the site, volume, and vascularity of the tumor, which can also be responsible for the large variations frequently observed in ctDNA levels[27]. Analysis of ctDNA can be performed by either quantitative assessment of ctDNA in a blood sample or by the identification of mutations. The introduction of next-generation sequencing (NGS)-based technologies reduced the error rate and enhanced sensitivity in ctDNA detection [28]. NGS technology enables the analysis of thousands of DNA sequences in parallel followed by either sequence alignment to a reference genome or de novo sequence assembly[29][30]. Deep sequencing represents the first approach to identify mutations at a low allele frequency (<0.2%) by sequencing the target regions with high coverage (>10,000×) [31]. Therefore, the sensitivity of deep sequencing for detecting mutations in ctDNA can achieve 100%, even if the specificity can be lower, around 80% [32]. Advantages of NGS included detection of genomic rearrangements, new mutations or alterations in genes, and the possible evaluation of response to treatment[33]. However, NGS-based approaches are rather expensive and time-consuming. Furthermore, data should be analyzed and interpreted by experts in bioinformatics[34]. Data storage and the difficulty in interpreting massive quantity of information obtained with NGS may represent a computational challenge to researchers. Also, the selection of proper validation methods to detect clinically significant mutations among a large number of samples can represent a challenging task[34]. Clinical validation of NGS data is carried out by assessing various parameters such as analytical sensitivity (the ability of the test to identify true sequence variants e.g., false negative rate), and analytical specificity (the probability of the test to not identify mutations where none are present (e.g., false positive rate)[35]. Limitations of NGS, principally with regard to the overall clinical sensitivity, could be overtaken implementing NGS with mutant allele enrichment or using digital PCR to improve reliability[36]. Mass-spectrometry and Real-Time PCR are other promising techniques for ctDNA assessment, which are rapid and cheap, require small quantities of input material, and have high sensitivity and specificity[35]. If possible, ctDNA should be analyzed in combination with CTCs and exosomal miRNAs, to obtain as much data as possible from a single blood sample[37]. However, different blood collection tubes, changes in storage temperatures and centrifugation may affect DNA or cells stability [38][39][40]. ctDNA degradation due to DNase activity could be avoided by isolating plasma within an hour after blood draw [41]. Reduction of cell lysis and stabilization of the total ctDNA pool can be obtained by means of specific blood collection tubes containing preservatives and additives[42]. Furthermore, accuracy and reproducibility of the liquid biopsy represent a main issue for analytic validity[43]. A study by Vivancos et al. showed that two liquid biopsy platforms, OncoBEAM™ RAS CRC and Idylla™ ctKRAS Mutation Test, had different sensitivity for identifying KRAS mutations in plasma samples from mCRC patients. The European Molecular Genetics Quality Network (EMQN) evaluated ctDNA detection approaches, and underlined that multiple pre-analytical and analytical variants may produce variable results; the EMQN pilot external quality assessment (EQA) scheme showed that the existing variability in multiple phases of ctDNA processing and analysis (e.g., due to specimen volume, ctDNA quantification technique, and choice of genotyping platform), resulted in an overall error rate of 6.09%[44]. These results highlighted the critical need for better standardization and validation of liquid biopsy assessment[47].
References
- GBD 2017 Colorectal Cancer Collaborators. The global, regional, and national burden of colorectal cancer and its attributable risk factors in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2019, 4, 913–933.
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424.
- Bishehsari, F.; Mahdavinia, M.; Vacca, M.; Malekzadeh, R.; Mariani-Costantini, R. Epidemiological transition of colorectal cancer in developing countries: Environmental factors, molecular pathways, and opportunities for prevention. World J. Gastroenterol. 2014, 20, 6055–6072.
- Hui, L. Quantifying the effects of aging and urbanization on major gastrointestinal diseases to guide preventative strategies. BMC Gastroenterol. 2018, 18.
- Valli, A.; Harris, A.L.; Kessler, B.M. Hypoxia metabolism in ageing. Aging Albany N. Y. 2015, 7, 465–466.
- Cancer Facts & Figures 2020|American Cancer Society. Available online: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2020.html (accessed on 14 July 2020).
- Stintzing, S. Management of colorectal cancer. F1000Prime Rep. 2014, 6, 108.
- Nakayama, G.; Tanaka, C.; Kodera, Y. Current Options for the Diagnosis, Staging and Therapeutic Management of Colorectal Cancer. Gastrointest. Tumors 2013, 1, 25–32.
- Bender, U.; Rho, Y.S.; Barrera, I.; Aghajanyan, S.; Acoba, J.; Kavan, P. Adjuvant therapy for stages II and III colon cancer: Risk stratification, treatment duration, and future directions. Curr. Oncol. 2019, 26, S43–S52.
- Pfeiffer, P.; Köhne, C.-H.; Qvortrup, C. The changing face of treatment for metastatic colorectal cancer. Expert Rev. Anticancer Ther. 2019, 19, 61–70.
- Moriarity, A.; O’Sullivan, J.; Kennedy, J.; Mehigan, B.; McCormick, P. Current targeted therapies in the treatment of advanced colorectal cancer: A review. Ther. Adv. Med. Oncol. 2016, 8, 276–293.
- Norcic, G. Liquid biopsy in colorectal cancer-current status and potential clinical applications. Micromachines 2018, 9, 300.
- Vacante, M.; Borzì, A.M.; Basile, F.; Biondi, A. Biomarkers in colorectal cancer: Current clinical utility and future perspectives. World J. Clin. Cases 2018, 6, 869–881.
- Palmirotta, R.; Lovero, D.; Cafforio, P.; Felici, C.; Mannavola, F.; Pellè, E.; Quaresmini, D.; Tucci, M.; Silvestris, F. Liquid biopsy of cancer: A multimodal diagnostic tool in clinical oncology. Ther. Adv. Med. Oncol. 2018, 10.
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484.
- Fernández-Lázaro, D.; García Hernández, J.L.; García, A.C.; Córdova Martínez, A.; Mielgo-Ayuso, J.; Cruz-Hernández, J.J. Liquid biopsy as novel tool in precision medicine: Origins, properties, identification and clinical perspective of cancer’s biomarkers. Diagnostics 2020, 10, 215.
- Carretero-González, A.; Otero, I.; Carril-Ajuria, L.; de Velasco, G.; Manso, L. Exosomes: Definition, role in tumor development and clinical implications. Cancer Microenviron. 2018, 11, 13–21.
- Wong, C.-H.; Chen, Y.-C. Clinical significance of exosomes as potential biomarkers in cancer. World J. Clin. Cases 2019, 7, 171–190.
- Beije, N.; Martens, J.W.M.; Sleijfer, S. Incorporating liquid biopsies into treatment decision-making: Obstacles and possibilities. Drug Discov. Today 2019, 24, 1715–1719.
- Kolenčík, D.; Shishido, S.N.; Pitule, P.; Mason, J.; Hicks, J.; Kuhn, P. Liquid Biopsy in Colorectal Carcinoma: Clinical Applications and Challenges. Cancers 2020, 12, 1376.
- Leers, M.P.G. Circulating tumor DNA and their added value in molecular oncology. Clin. Chem. Lab. Med. CCLM 2020, 58, 152–161.
- Hong, B.; Zu, Y. Detecting circulating tumor cells: Current challenges and new trends. Theranostics 2013, 3, 377–394.
- Mamdouhi, T.; Twomey, J.D.; McSweeney, K.M.; Zhang, B. Fugitives on the run: Circulating tumor cells (CTCs) in metastatic diseases. Cancer Metastasis Rev. 2019, 38, 297–305.
- Millner, L.M.; Linder, M.W.; Valdes, R. Circulating tumor cells: A review of present methods and the need to identify heterogeneous phenotypes. Ann. Clin. Lab. Sci. 2013, 43, 295–304.
- Barrière, G.; Tartary, M.; Rigaud, M. Epithelial mesenchymal transition: A new insight into the detection of circulating tumor cells. ISRN Oncol. 2012, 2012.
- Castro-Giner, F.; Gkountela, S.; Donato, C.; Alborelli, I.; Quagliata, L.; Ng, C.K.Y.; Piscuoglio, S.; Aceto, N. Cancer diagnosis using a liquid biopsy: Challenges and expectations. Diagnostics 2018, 8, 31.
- Elazezy, M.; Joosse, S.A. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput. Struct. Biotechnol. J. 2018, 16, 370–378.
- D’Haene, N.; Fontanges, Q.; De Nève, N.; Blanchard, O.; Melendez, B.; Delos, M.; Dehou, M.-F.; Maris, C.; Nagy, N.; Rousseau, E.; et al. Clinical application of targeted next-generation sequencing for colorectal cancer patients: A multicentric Belgian experience. Oncotarget 2018, 9, 20761–20768.
- Qin, D. Next-generation sequencing and its clinical application. Cancer Biol. Med. 2019, 16, 4–10.
- Narayan, A.; Carriero, N.J.; Gettinger, S.N.; Kluytenaar, J.; Kozak, K.R.; Yock, T.I.; Muscato, N.E.; Ugarelli, P.; Decker, R.H.; Patel, A.A. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res. 2012, 72, 3492–3498.
- Couraud, S.; Vaca-Paniagua, F.; Villar, S.; Oliver, J.; Schuster, T.; Blanché, H.; Girard, N.; Trédaniel, J.; Guilleminault, L.; Gervais, R.; et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: A proof-of-concept study from BioCAST/IFCT-1002. Clin. Cancer Res. 2014, 20, 4613–4624.
- Fontanges, Q.; De Mendonca, R.; Salmon, I.; Le Mercier, M.; D’Haene, N. Clinical application of targeted next generation sequencing for colorectal cancers. Int. J. Mol. Sci. 2016, 17, 2117.
- Grada, A.; Weinbrecht, K. Next-generation sequencing: Methodology and application. J. Investig. Dermatol. 2013, 133, e11.
- Ng, C.; Li, H.; Wu, W.K.K.; Wong, S.H.; Yu, J. Genomics and metagenomics of colorectal cancer. J. Gastrointest. Oncol. 2019, 10, 1164–1170.
- Kim, R.Y.; Xu, H.; Myllykangas, S.; Ji, H. Genetic-based biomarkers and next-generation sequencing: The future of personalized care in colorectal cancer. Pers. Med. 2011, 8, 331–345.
- Wadapurkar, R.M.; Vyas, R. Computational analysis of next generation sequencing data and its applications in clinical oncology. Inform. Med. Unlocked 2018, 11, 75–82.
- Rachiglio, A.M.; Abate, R.E.; Sacco, A.; Pasquale, R.; Fenizia, F.; Lambiase, M.; Morabito, A.; Montanino, A.; Rocco, G.; Romano, C.; et al. Limits and potential of targeted sequencing analysis of liquid biopsy in patients with lung and colon carcinoma. Oncotarget 2016, 7, 66595–66605.
- Del Vecchio, F.; Mastroiaco, V.; Di Marco, A.; Compagnoni, C.; Capece, D.; Zazzeroni, F.; Capalbo, C.; Alesse, E.; Tessitore, A. Next-generation sequencing: Recent applications to the analysis of colorectal cancer. J. Transl. Med. 2017, 15, 246.
- Soda, N.; Rehm, B.H.A.; Sonar, P.; Nguyen, N.-T.; Shiddiky, M.J.A. Advanced liquid biopsy technologies for circulating biomarker detection. J. Mater. Chem. B 2019, 7, 6670–6704.
- Risberg, B.; Tsui, D.W.Y.; Biggs, H.; Ruiz-Valdepenas Martin de Almagro, A.; Dawson, S.-J.; Hodgkin, C.; Jones, L.; Parkinson, C.; Piskorz, A.; Marass, F.; et al. Effects of Collection and Processing Procedures on Plasma Circulating Cell-Free DNA from Cancer Patients. J. Mol. Diagn. 2018, 20, 883–892.
- Grölz, D.; Hauch, S.; Schlumpberger, M.; Guenther, K.; Voss, T.; Sprenger-Haussels, M.; Oelmüller, U. Liquid Biopsy Preservation Solutions for Standardized Pre-Analytical Workflows—Venous Whole Blood and Plasma. Curr. Pathobiol. Rep. 2018, 6, 275–286.
- Harouaka, R.; Kang, Z.; Zheng, S.; Cao, L. Circulating tumor cells: Advances in isolation and analysis, and challenges for clinical applications. Pharmacol. Ther. 2014, 141, 209–221.
- Tamkovich, S.N.; Cherepanova, A.V.; Kolesnikova, E.V.; Rykova, E.Y.; Pyshnyi, D.V.; Vlassov, V.V.; Laktionov, P.P. Circulating DNA and DNase activity in human blood. Ann. N. Y. Acad. Sci. 2006, 1075, 191–196.
- Siravegna, G.; Mussolin, B.; Venesio, T.; Marsoni, S.; Seoane, J.; Dive, C.; Papadopoulos, N.; Kopetz, S.; Corcoran, R.B.; Siu, L.L.; et al. How liquid biopsies can change clinical practice in oncology. Ann. Oncol. 2019, 30, 1580–1590.
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88.
- Haselmann, V.; Ahmad-Nejad, P.; Geilenkeuser, W.J.; Duda, A.; Gabor, M.; Eichner, R.; Patton, S.; Neumaier, M. Results of the first external quality assessment scheme (EQA) for isolation and analysis of circulating tumour DNA (ctDNA). Clin. Chem. Lab. Med. 2018, 56, 220–228.
- Vivancos, A.; Aranda, E.; Benavides, M.; Élez, E.; Gómez-España, M.A.; Toledano, M.; Alvarez, M.; Parrado, M.R.C.; García-Barberán, V.; Diaz-Rubio, E. Comparison of the Clinical Sensitivity of the Idylla Platform and the OncoBEAM RAS CRC Assay for KRAS Mutation Detection in Liquid Biopsy Samples. Sci. Rep. 2019, 9.




