Arginine availability and activation of arginine-related pathways at cancer sites have profound effects on the tumor microenvironment, far beyond their well-known role in the hepatic urea cycle. Arginine metabolism impacts not only malignant cells but also the surrounding immune cells behavior, modulating growth, survival, and immunosurveillance mechanisms, either through an arginase-mediated effect on polyamines and proline synthesis, or by the arginine/nitric oxide pathway in tumor cells, antitumor T-cells, myeloid-derived suppressor cells, and macrophages.
1. Arginine Metabolism
Arginine, the substrate of arginase, is a nonessential cationic amino acid obtained from diet, endogenous synthesis, and protein turnover and is essential for protein synthesis and a precursor of several molecules such as urea, nitric oxide, polyamines, proline, and agmatine, among others [1] (Figure 1).
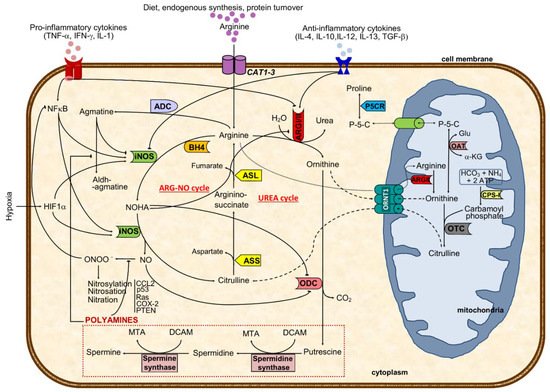
Figure 1. Major cellular arginine metabolic routes and regulatory mechanisms. The aminoacid arginine is transported to the cell by CAT transporters or synthesized from citrulline. Intracellular arginine can either be metabolized by arginase I or II (ARGI or ARGII) to ornithine and urea (excreted out of the cell and of the body) or to NO and citrulline through iNOS catalysis, depending on iNOS/ARG ratio. ARGI expression and activity are regulated positively by anti-inflammatory and negatively via pro-inflammatory cytokines and NOHA, an intermediate in NO formation. In turn, iNOS activity is negatively regulated by agmatine, a decarboxylated product of arginine, and by anti-inflammatory cytokines, besides the positive pro-inflammatory and hypoxic stimulus. Preferential production of ornithine through ARG catalysis can either result in activation of the polyamines pathway (putrescine, spermidine, and spermine) or in ornithine shuttling to the mitochondria (where it might follow the pathway towards proline formation or be transformed in citrulline and shuttled back to the cytoplasm). Arginine antiported to the mitochondria will increase the ornithine pool after ARGII catalysis. When iNOS metabolizes arginine, the resulting products are citrulline and nitric oxide using BH4 as a co-factor. While NO might be transformed to peroxynitrite that may nytrosylate/nitrosate/nitrate key proteins to become pro-tumoral, citrulline formation will drive the ARG-NO recycling. Citrulline from UREA or ARG-NO cycles is converted to arginine-succinate (by ASS), which will result in arginine and fumarate (catalyzed by ASL). This last step represents a link with the Krebs cycle. Solid black lines with arrows indicate a positive effect, whereas solid lines with blunted ends specify an inhibitory effect. Dashed line indicates ornithine-citrulline antiport through mitochondrial membrane, whereas dotted line represents transportation of arginine into the mitochondria or transportation of P-5-C between cytoplasm and mitochondria. ASL, argininosuccinate lyase; ASS, argininosuccinate synthase; CPS-I, carbamoyl phosphate synthetase I; OAT, ornithine aminotransferase; ARG, arginase (type I and type II); ARG-NO cycle, arginine-nitric oxide cycle where iNOS is primarily involved in arginine metabolism; ADC, arginine decarboxylase; ODC, ornithine decarboxylase; OTC, ornithine transcarbamylase; iNOS, inducible nitric oxide synthase; L-HydroxyArg, L-hydroxyarginine; NO, nitric oxide; P-5-C, pyrroline-5-carboxylate; P5CR, pyrroline-5-carboxylate reductase; CAT1-3, cationic amino acid transporters (CAT1 is found in epithelial cells and CAT2 in macrophages); CCL2, Chemokine (C-C motif) ligand 2 or monocyte-chemoattractant protein 1; HIF1α, hypoxia-inducible factor 1 α; NFκB, nuclear factor κB; ORNT1, mitochondrial ornithine: citrulline antiporter; BH4, tetrahydrobiopterin; DCAM, decarboxylated 5-adenonosylmethionine; Glu, glutamate; MTA, methylthioadenosine; NOHA, N-hydroxy-L-arginine; α-KG, α-ketoglutamate; p53, tumor protein 53; Ras, Ras oncogene; COX-2, cyclooxygenase 2; PTEN, phosphatase and tensin homolog; UREA cycle, urea cycle where ARGI is primarily involved in arginine metabolism.
Interestingly, while minor arginine disturbances might prompt cellular and organ dysfunctions, its metabolism is highly intricate and tightly regulated. It is synthesized in the cytoplasm from citrulline through consecutive reactions catalyzed by argininosuccinate synthase (ASS) and argininosuccinate lyase (ASL) or transported to the cell by cationic amino acid transporters (CATs 1-3) (
Figure 1). The combination of CATs compartmentalization with the different isoforms of nitric oxide synthases has been recently reviewed
[2].
Then, in the urea cycle (mainly at the liver), arginase converts arginine into urea and ornithine
[3][4]. Hereafter, ornithine can either be recycled again into arginine through the urea cycle or converted to polyamines (by ornithine decarboxylase, ODC) or shuttled to the mitochondria where it is used for proline synthesis (via ornithine aminotransferase) (
Figure 1)
[5][6]. The rate-limiting enzyme ODC, the first in the biosynthesis of the polyamine that converts ornithine into putrescine, has been shown to facilitate tumorigenesis and invasiveness, being highly expressed in tumors
[5][7]. In mitochondria, ornithine is catalyzed by ornithine aminotransferase to pyrroline–5-carboxylate, which is shuttled back to cytosol and reduced to proline (
Figure 1). The pyrroline–5-carboxylate-to–proline conversion can transfer the redox potential across the mitochondrial membrane and is involved in several processes, such as protein phosphorylation, platelet activation, cell signaling, hypoxia, cell growth control, and collagen production, which are all relevant in cancer
[6][8].
2. Arginine Metabolism and Cancer
High arginase activity has been described in different malignancies, including of the skin, colon, breast, hematologic, and prostate, mainly due to their need to produce polyamines and respond to rapid proliferation
[9][10][11][12][13][14]. In agreement, tumor cell lines (some resistant to ODC inhibitors) holding arginase activity resulted in lower production of polyamines, reduced cell proliferation, and induction of apoptosis after inhibition of ARG activity
[15][16].
Besides the mechanism of substrate deprivation for cancer growth, in which arginases and arginine availability have a nearly direct effect in cancer cells, current perspectives elicited an extended explanation focused on the interplay of arginases metabolism and arginine among components of the tumor microenvironment as mediators of immunosurveillance. Notably, the efficacy of chimeric antigen receptor T cells (CAR-T) in hematological and solid malignancies has been shown to be compromised by depletion of arginine due to the low expression of arginine resynthesis enzymes, ASS, and ornithine transcarbamylase; therefore, the re-engineering of T cells may correct the mentioned defect and induce the expression of functional ASS or ornithine transcarbamylase enzymes, increasing CAR-T cell proliferation without compromising function
[17].
Arginine depletion exerted target transcriptional and posttranscriptional regulation of ASS and ASL activities and regulation of other genes such as those coding for iNOS, CAT-1 or T-cell receptor (TCR) zeta-chain, thereby modulating relevant cellular mechanisms (
Figure 2): (i) there is a diminished efficiency in iNOS mRNA translation and protein stability triggered by lower extracellular arginine and arginases overactivity due to the downregulation of eukaryotic initiation factor 2α (eIF2α)
[5][18][19]; (ii) the lower TCR zeta-chain mRNA stability and expression affects lymphocytes proliferative capability
[20][21]; (iii) both transcriptional and translational efficiency of CAT-1 mRNA are increased
[22][20], conferring advantage for capturing extracellular arginine to tumor cells.
During tumor progression, macrophage polarization switches from classically activated M1, which promotes tumor initiation and adaptive immunity activation towards an alternatively activated (M2) polarized state, favoring tumor progression and spread
[23]. In M1, inflammatory mediators regulate ASS expression together with iNOS, ASL, and CAT-2 activities, thus providing increased arginine synthesis and uptake for NO production. This regulation of arginine and NO by inflammatory mediators is also found in other cell types (e.g., epithelial cells and vascular smooth muscle) and establishes the arginine-NO pathway that supports NO generation (
Figure 1)
[6][22]. M1 macrophages may contribute to the eradication of cancer cells once they generate NO, pro-inflammatory cytokines, and chemokines, or may act as antigen-presenting cells to activate CD8+ cytotoxic T-cells
[24]. Conversely, M2 macrophages promote tumorigenesis through anti-inflammatory cytokines and chemokines (IL-4, IL-13, and transforming growth factor β, TGF-β pathways)
[24] (
Figure 2).
In macrophages, both ARGI and ARGII and NOS expression and activity were identified
[25][26]. ARGI is markedly induced in human mononuclear cells after tissue injury, associating with decreases in arginine and NO availability
[26][27]. ARGI is inducible, while ARGII is constitutively expressed in macrophages and tightly regulated by Th1 and Th2 cytokines that modulate the production of ornithine and subsequent products
[28][26]. Indeed, pro-inflammatory cytokines (tumor necrosis factor alpha, TNF-α; interleukin 1, IL-1; interferon gamma, IFN-γ) induce iNOS and suppress arginase activity, whereas anti-inflammatory cytokines (IL-4, IL-12, IL-13, and IL-10) reduce iNOS and increase arginase activity
[5][8][28] (
Figure 2). As in epithelial cells, also in macrophages, the synthesis of NO is dependent on arginase expression, which is well correlated with arginase activity
[8][27][29].
The key inflammation-related transcription factors nuclear factor erythroid 2-like 2 and nuclear factor kappa-B can be modulated by NO in malignant cells promoting cell survival and tumor progression
[30]. Even though tumor cell-induced iNOS is involved in those processes through NO production, its activation in macrophages further increases the inflammatory burden
[31][32]. Additionally, arginase induction in macrophages decreases NO production by conversion of arginine in ornithine
[33][31][27][34][35]. In this process, ARGII and ODC are initially induced and may lead to increased polyamine production
[26]; and then followed by iNOS downregulation, further relocating arginine to ornithine synthesis that can be used for polyamines and proline synthesis, essential for cell proliferation and tissue remodeling
[5][8][26][27][36].
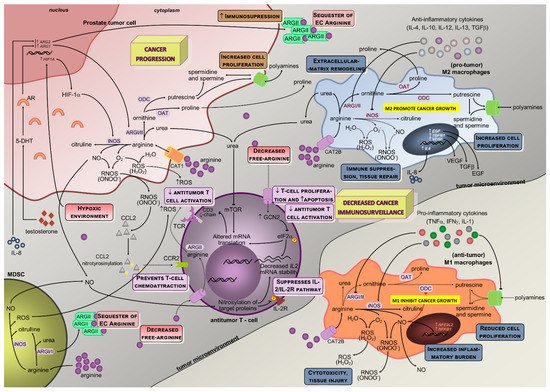
Figure 2. Schematic representation of arginine metabolism and the interplay between cellular players in the prostate tumor microenvironment. The activities of ARGs and iNOS are illustrated, together with arginine-activated downstream pathways in cellular components of the tumor microenvironment (androgen-responsive prostate cancer cell, macrophages, antitumor T-cell, MSCs). The most relevant pathophysiological implications of arginine metabolism are reduced cancer immunosurveillance and a stimulatory action in prostate malignant cells towards cancer progression. Solid black lines with arrows indicate the main enzymatic activity or movement of molecules, whereas dashed lines indicate alternative metabolic pathways or suppression of the movement of molecules. Solid blue lines with arrows designate a stimulatory effect in enzyme activity, while solid red lines with blunt ends specify inhibition of enzyme activity or transporter activity. While pro-tumoral M2 macrophages present increased activity of ARGs with subsequent proline and polyamines production that result in collagen deposition and higher cell proliferation, the antitumoral M1 macrophages (commonly found out of the tumor microenvironment) have an overactive iNOS pathway with resulting pro-inflammatory stimulus (NRF2L2 and NFKB1 overexpression), reduced cell growth and production of reactive oxygen (hydrogen peroxide) and nitrogen species (peroxynitrites) that ultimately induce cytotoxicity and tissue injury. The iNOS and ARGs enzymes are tightly regulated by cytokine and metabolic circuits, although these enzymes also directly activate biochemical circuits that negatively regulate each other. As a resulting effect of arginine metabolism and capture in M2 macrophages, several pro-tumoral growth factors are produced and secreted to the tumor microenvironment (VEGF, EGF, TGFβ), while reducing the extracellular pool of free arginine that will contribute to their immune suppression and tissue repair phenotype. By producing IL-4, IL-10, and TGF-β1 anti-inflammatory cytokines, tumor cells also contribute not only to macrophages differentiation towards M2 but also to the regulation of arginine metabolism in macrophages (activating ARGs and downregulating iNOS). Together with tumor cells and MDSCs, macrophages also contribute to increased levels of urea as a result of higher ARGs activity in these cells, which will impair mTOR signaling and T-cell receptor CD3 ζ-chain subunit mRNA translation, thus resulting in hindered antitumor T-cell activation. The hypoxic tumoral microenvironment increases HIF1A expression and protein production by tumor cells, which signals downstream to increase arginine-iNOS pathway activation that results in increased production of nitric oxide, and reactive nitrogen and oxygen species. While NO can impact antitumor T-cell activation through nitrosylation of target proteins and suppression of the IL-2/IL-2R pathway, peroxynitrites may hamper CCL2 binding and prevent T-cell chemoattraction towards tumors, and reactive oxygen species can influence negatively antitumor T-cell activation. Moreover, the particularity of prostate tumor cells’ dependence on androgens implies its influence in ARG1 and ARG2 expression, which will potentiate the urea cycle towards polyamines and proline production with resulting increases in cell proliferation and collagen synthesis. In addition, the overexpression of ARG1 and ARG2 will lead to ARGs exportation out of the cell, where they might sequester extracellular arginine, further increasing immunosuppression. MDSCs also metabolize arginine either through the ARG-NO cycle or the UREA cycle (Figure 1). MDSCs in the tumor microenvironment might secrete NO, reactive oxygen species, and ARGs receptors out of the cell, which will contribute to the suppression of IL-2/IL-2R pathway of T-cells, decreased activation of antitumor T-cells and sequestering of extracellular arginine, respectively, ultimately leading to reduced cancer immunosurveillance. 5-DHT, 5alpha-dihydrotestosterone; AR, androgen receptor; ARG1, gene coding for the arginase type I; ARG2, gene coding for the arginase type II; CAT, cationic amino acid transporter; CCL2, chemokine-CC motif-ligand 2; CCR2, chemokine-CC motif-receptor 2; EGF, epidermal growth factor; eIF2α, eukaryotic initiation factor 2α; GCN2, general control of nutrition; HIF-1α, hypoxia inducible factor 1 alpha; HIF1A, gene coding for the hypoxia inducible factor 1 alpha; IFNγ, interferon γ; IL-1, interleukine 1; IL-10, interleukine 10; IL-12, interleukine 12; IL-13, interleukine 13; IL2, gene coding for the interleukine 2; IL-2, interleukine 2; IL-2R, interleukine 2 receptor; IL-4, interleukine 4; IL-8, interleukine 8; iNOS, inducible nitric oxide synthase; MDSC, myeloid-derived suppressor cells; mTOR, mammalian target of rapamycin; NFE2L2, gene coding for the nuclear factor erythroid 2-like 2 (Nrf2); NFKB1, gene coding for the nuclear factor kappa-b subunit 1; NO, nitric oxide; OAT, ornithine aminotransferase; ARG, arginase (type I and type II); ODC, ornithine decarboxylase; RNOS, reactive nitrogen species; ROS, reactive oxygen species; TCR CD3 ζ-chain, CD3 ζ-chain in T cell receptor; TCR, T-cell receptor; TGFβ, transforming growth factor beta; TNFα, tumoral necrosis factor α; VEGF, vascular endothelial growth factor.
The lower availability of arginine in the microenvironment that results from upregulated arginine metabolism in tumor cells, in M2 macrophages, and in myeloid-derived suppressor cells (MDSCs), exerts a regulatory effect in lymphocyte activation through downregulation of the TCR CD3ζ chain, central for signaling and responsiveness in activated T cells
[37]. It was shown that arginase upregulation, together with CAT-2 transporter under expression in tumor-surrounding cells, reduces arginine availability and downregulates CD3ζ T cell receptor expression
[20][26][37], with an inhibitory impact on antitumor T cell activity. This mechanism might be used by anti-inflammatory macrophages, M2-like, to modulate T-cell function in the tumor microenvironment
[21]. Taken together, these results suggest that decreased arginine availability and increased arginine metabolism in cells adjacent to lymphocytes may have a suppressive role in the cell-mediated immune response to cancer.
3. Modulation of Arginine Metabolism in Oncology
Modulation of arginine availability and arginase activity by targeting the enzymes in its metabolic pathway may have a role in cancer therapeutics. The auxotrophic affinity of cancer cells for specific amino acids has been the rationale for therapeutic deprivation regimens, where arginine fits, once it is required by proliferating cells, despite nonessential to normal cells. Downregulation of the rate-limiting arginine-producer enzyme ASS in tumor cells, which is common in most cancers [38], correlates with the dependence on extracellular arginine due to the inability to produce endogenous arginine for growth [15]. Thus, arginine-depleting enzymes involved in arginine catabolism out of the cell (arginase and arginine deiminase, androgen-independent, a microbial enzyme that converts arginine to citrulline and ammonia) may have an antitumor effect, with tumoral ASS deficiency serving as a prognostic biomarker and predictor of sensitivity to arginine deprivation therapy [16].
Citrullination is a deimination of protein-embedded arginine, which is converted to the non-coded amino acid citrulline [39]. This process is catalyzed by a family of enzymes called peptidyl arginine deiminases. Citrullination is involved in disease pathogenesis with the involvement of the following mechanisms epigenetic, pluripotency, immunity, and transcriptional regulation. Indeed, peptidyl arginine deiminase 2-mediated arginine citrullination might have an implication on transcriptional regulation in cancer [40]. Furthermore, citrullination of histones in neutrophils facilitates neutrophil extracellular trap formation or NETosis [41].
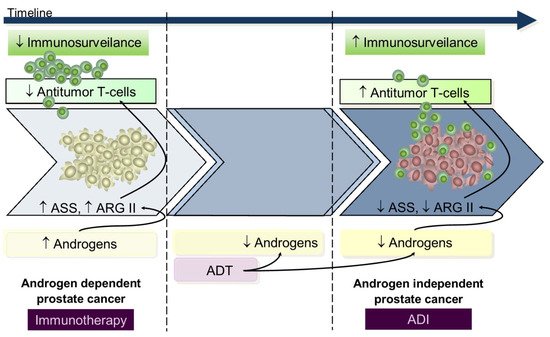
Data from in vitro and in vivo studies using either in androgen-dependent or androgen-independent prostate cancer cell lines suggests that during the androgen-dependent phase, with higher ASS and ARGII expression, tumors were resistant to androgen-independent, whereas in the androgen-independent stage, presenting decreased ASS and ARGII expression, there was a response to androgen-independent treatment [9][42]. From this perspective, we hypothesize that during the early androgen-dependent PCa development preeminent mechanisms contribute to immunosuppression rendering immunotherapy a key role; on the contrary, advanced androgen-independent tumors that survive in low androgen environments have increased T-cells infiltrated [43] and low ASS and ARGII expression [9][42] rendering androgen-independent a potential therapeutic utility since lymphocyte activity is already restored (Figure 3); however, despite these findings from in vitro and animal models [15][44][45][46], and in patients with metastatic cancer [47][48][49], the apparently low potency of ARG and AD androgen-independent immunogenicity advise restrained enthusiasm.
Figure 3. Arginine metabolism according to prostate cancer hormonal status informs plausible therapies. During early AD PCa development, androgens are available and contribute to increased immunosuppression and proliferative stimulus through ARGII mediation. In this setting, increased ASS activity confers resistance to ADI administration, and the most benefit is likely to derive from immunotherapy. Conversely, emergence of the androgen-independent phase of disease, after acquired resistance to ADT therapy, is associated with tumor cell survival in very low androgen levels with subsequent low ASS and ARGII expression and normal immune activity (conferring higher response rate to ADI treatment). ADT, androgen deprivation therapy; ADI, arginine deiminase; ARGII, arginase II; ASS, argininosuccinate synthase.