Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Tao Wang | + 2174 word(s) | 2174 | 2021-05-18 05:04:35 | | | |
| 2 | Catherine Yang | -21 word(s) | 2153 | 2021-06-01 11:33:11 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Wang, T. Taxol Biosynthetic Pathway. Encyclopedia. Available online: https://encyclopedia.pub/entry/10352 (accessed on 08 February 2026).
Wang T. Taxol Biosynthetic Pathway. Encyclopedia. Available at: https://encyclopedia.pub/entry/10352. Accessed February 08, 2026.
Wang, Tao. "Taxol Biosynthetic Pathway" Encyclopedia, https://encyclopedia.pub/entry/10352 (accessed February 08, 2026).
Wang, T. (2021, June 01). Taxol Biosynthetic Pathway. In Encyclopedia. https://encyclopedia.pub/entry/10352
Wang, Tao. "Taxol Biosynthetic Pathway." Encyclopedia. Web. 01 June, 2021.
Copy Citation
Taxol is one of the most effective anticancer drugs in the world that is widely used in the treatment of several cancers. The elucidation of the taxol biosynthetic pathway is the key to solve the problem of taxol supply. So far, the taxol biosynthetic pathway has been reported to require an estimated 20 steps of enzymatic reactions, and most of enzymes involved have been well characterized. In details, the source and formation of the taxane core and the process of the downstream synthetic pathway have been basically depicted, while the modification of the core taxane skeleton has not been fully reported, mainly concerning the developments from diol intermediates to 2-debenzoyltaxane.
taxol
biosynthetic pathway
latest research progress
1. Introduction
Taxol, a kind of tetracyclic diterpenoid secondary metabolite originally extracted from the bark of Taxus brevifolia [1], is recognized as one of the most effective antitumor drugs around the world. Taxol could inhibit the division of tumor cells in the G2/M phase of the cell cycle by promoting the formation and stability of microtubules, where it was approved by the U.S. Food and Drug Administration (FDA) for the treatment of several cancers, such as breast, lung and ovarian cancer [2]. Recently, the annual sales of taxol and its related products have exceeded $1 billion [3]. However, the content of taxol in the Taxus species is extremely low (the highest content in the bark is only about 0.06%), and the slow growth rate, lack of resources and the poor competitiveness and regeneration ability in the Taxus population, resulting in the insufficient yield of natural harvest to meet the growing market demand [4].
Several alternative methods have been explored to increase taxol production. The chemical total synthesis of taxol was reported by Holton et al. [5] and Nicolaou et al. [6] simultaneously with different synthetic routes, but neither of them was commercially viable due to the complex routes, uncontrollable reaction conditions and the low yields [3][7]. The semisynthetic method involves extracting relatively high levels of taxol precursor baccatin III or 10-deacetyl baccatin III (10-DAB) from the reproducible twigs and leaves of cultivated Taxus species, and then converting to taxol by chemical synthesis. This is so far the main approach for industrial production of taxol, but it relies heavily on Taxus resources in essence [8][9]. Moreover, the extractions of taxol from Taxus cell culture and taxol-producing endophytic fungi are also expected, because they are controllable and environmentally sustainable, but no major breakthrough has been achieved at this stage [10][11][12].
Recently, a series of encouraging progress has been made in the production of taxol through heterologous synthesis. Ajikumar et al. [13] reported a multivariate modular pathway engineering, which increased the yield of taxadiene, the first committed taxol intermediate, to approximately 1 g/L in an engineered Escherichia coli strain; the yield of taxadien-5α-ol was also up to 58 ± 3 mg/L. Stable coculture of Saccharomyces cerevisiae and E. coli in the same bioreactor was reported by Zhou et al. [14], who designed specialized environments or compartments for optimal function, and produced 33 mg/L oxygenated taxanes, including a monoacetylated dioxygenated taxane, the taxadien-5a-acetate-10β-ol. Moreover, several plant hosts have also been exploited to produce taxol intermediates, such as Arabidopsis thaliana [15], tomato [16] and Physcomitrella patens [17]. However, the accumulation of taxadiene could cause the growth retardation and decreased levels of photosynthetic pigment in transgenic plants. Fortunately, Li et al. [18] used a chloroplastic compartmentalized metabolic engineering strategy, which produced taxadiene and taxadiene-5α-ol at 56.6 μg/g FW and 1.3 μg/g FW, respectively, in a high biomass plant Nicotiana benthamiana. This study highlights the potential of tobacco as an alternative platform for taxol production. Production of taxol and its precursors in heterologous hosts is more sustainable than extraction from tissues of Taxus trees or chemical synthesis. However, it still cannot circumvent the bottlenecks in the taxol biosynthetic pathway. Further developments from diol intermediates to functional taxanes are rarely reported.
There is no doubt that the synthesis and yield of taxol will continue to depend on the improvement of biological methods for the foreseeable future. In view of this, it is essential to explore alternative methods and increase the taxol yield, which mainly depends on the understanding of the taxol biosynthetic pathway in detail.
2. Taxol Biosynthetic Pathway
The taxol biosynthetic pathway has been basically elucidated, which requires estimated 20 enzymatic steps from the common diterpenoid precursor geranylgeranyl diphosphate (GGPP) to taxol itself. As shown in Figure 1, this complex process can be divided into three parts: the source and formation of taxane core, the modification of the core taxane skeleton and the synthesis of β-phenylalanyl-CoA side chain and the assembly of taxol.
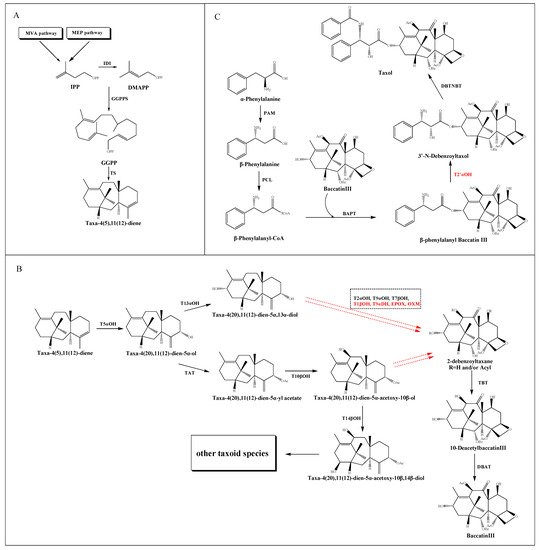
Figure 1. Putative taxol biosynthetic pathway. (A) The source and formation of the taxane core. (B) The modification of the core taxane skeleton. (C) The synthesis of the β-phenylalanyl-CoA side chain and the assembly of taxol. The solid arrows show the identified steps. The dotted arrows show the unknown steps. Steps marked by red color represent the uncharacterized enzymatic reactions. Dotted frame represents intermediate steps of the pathway where several enzymes and the order of their reactions are unknown.
2.1. The Source and Formation of Taxane Core
The taxane core, namely taxadiene, derived from the cyclization of the GGPP catalyzed by taxadiene synthase (TS) [19]. In detail, GGPP is converted from three molecules of isopentenyl diphosphate (IPP) and one of dimethylallyl diphosphate (DMAPP) catalyzed by geranylgeranyl diphosphate synthase (GGPPS) [20]. IPP is considered as a common intermediate in the cytosolic mevalonic acid (MVA) pathway and the plastid 2-C-methyl-D-erythritol-4-phosphate (MEP) pathway. The conversion of IPP to DMAPP is catalyzed by a plastid IPP isomerase (IDI) [21]. Recently, the dispute over the source of IPP and DMAPP for the taxol pathway has made substantial progress. Despite evidence suggested that fosmidomycin and mevinolin could inhibit the plastid and cytosolic IPP pathways of T. baccata cells respectively [22][23], their selective blocking function in a TS-transgenic N. benthamiana plant suggesting that the MEP pathway supplies the bulk of the isoprenoid precursors for the taxane skeleton [18]. Moreover, the coexpression model of taxol synthesis related genes and upstream genes in T. cuspidate suggested a local IPP bias from the plastids [24] (Figure 1A).
2.2. The Modification of the Core Taxane Skeleton
After the synthesis of taxane skeleton, multiple tailoring enzymes are involved in modifying the skeleton at the C-1, C-2, C-4, C-5, C-7, C-9, C-10 and C-13 positions (Figure 1B). Firstly, a class II cytochrome P450 (CYP450) hydroxylase called taxadiene-5α- hydroxylase (T5αOH) [25] converts taxadiene into taxadien-5α-ol via hydroxylation at the C-5 position and migration of the carbon double bond. Then, the taxol pathway has been reported in two speculations: the taxadien-5α-ol is instigated by either a hydroxylation at the C-13 position to form taxadien-5α,13α-diol [26], or an acylation at C-5 position to form taxadien-5α-yl acetate [27], which could be further hydroxylated at C-10 position to form taxadien-5α-acetoxy-10β-ol [28]. Recently, a hydroxylation at the C-14 position leads to the formation of taxadien-5α-acetoxy-10β,14β-diol, but the intact structure of taxane core showed no modification at C-14 position, which implied a putative bifurcation leads to the biosynthesis of other taxoids [29]. The hydroxylation of the taxane skeleton has proven to be mediated by the CYP450 family. However, these CYP450s are selective for acetylated and polyoxygenated taxadiene substrates. Trials to produce triols from the diol intermediates using the corresponding CYP450s are currently unsuccessful.
The order of the catalytic steps after diol intermediates is not yet clear, including a series of hydroxylation at C-1, C-2, C-4, C-7 and C-9 positions, a further oxidation at C-9 position, and a formation of oxetane ring at C-4 and C-5 positions. Since the oxygenation of C-9 is believed to be an early event, while the oxygenation of C-1 happens later, Holton et al. [30] reported the order of hydroxylation should be C5, C10, C13, C9, C7, C2 and C1. However, according to the ratio of oxygenated taxoids found in Taxus cells, Vongpaseuth and Roberts [31] considered the order of C5, C10, C13, C2, C9, C7 and C1 may be more accurate. A series of transformations produce the hypothetical precursor 2-debenzoyltaxane, which could be further catalyzed by the enzyme taxane-2α-O-benzoyl transferase (TBT) to produce 10-DAB [32], one of the most important precursors for the chemical semisynthesis of taxol. Then a 10-deacetylbaccatin III-10-O-acetyl transferase (DBAT) could catalyze 10-DAB at the C-10 position to generate baccatin III, which is considered to be the last reaction in the formation of taxane core [33].
2.3. The Synthesis of β-Phenylalanyl-CoA Side Chain and the Assembly of Taxol
The β-phenylalanyl-CoA side chain is derived from α-phenylalanine, which is first converted into β-phenylalanine by phenylalanine aminomutase (PAM) [34]. Subsequently, a β-phenylalanyl-CoA ligase (PCL) [35] is predicted to convert β-phenylalanine into β-phenylalanyl-CoA. This intermediate is then attached to the C13 hydroxyl group of the taxane core by the enzyme baccatin III: 3-amino, 13-phenylpropanoyltransferase (BAPT) to produce β-phenylalanyl baccatin III [36], which is then hydroxylated at the C2’ position of the side chain, and terminal N-benzoylation at the C3’ position by the enzyme N-benzoyl transferase (DBTNBT) to produce the final product taxol [37] (Figure 1C).
2.4. Key Enzymes Involved in the Taxol Pathway
So far, sixteen enzymes involved in the taxol pathway have been well characterized (Table 1), including a novel C10 hydroxylase [25] and a putative PCL. The enzymes responsible for C-1 hydroxylation, C-2′ hydroxylation and C-9 oxidation are currently unknown, but are predicted to belong to the CYP450 family. Moreover, neither the proposed C4β, C20-epoxidase (EPOX) nor the oxomutase (OXM) responsible for the formation of oxetane ring has been identified, but potential gene candidates have been discovered through the analysis of jasmonate-induced T. baccata cultures [35]. Interestingly, both taxane-2α-hydroxylase (T2αOH) and taxane-7β-hydroxylase (T7βOH) could use taxusin as a substrate to form 2α-hydroxytaxusin and 7β-hydroxytaxusin, respectively. Then, these intermediates can reciprocally convert the corresponding hydroxyl products of the respective reactions to the common 2α,7β-dihydroxytaxusin [38][39] (Figure 2A). This suggested that the taxol biosynthetic pathway may not be a single linear, but a complex network of anastomosing routes that potentially have several common nodes. Moreover, the enzyme taxane-9α-hydroxylase (T9αOH) was characterized by Zhang et al. [40] in Ginkgo biloba cells, which can convert Sinenxan A (SIA) to form 9α-hydroxy-SIA (Figure 2B). This further implied that the process in the production of 2-debenzoyltaxane may involve transient acylation/deacylation for the purposes of trafficking and organellar targeting, or flux regulation. Such processes also greatly increase the number of biosynthetic steps and pathway complexity [41].
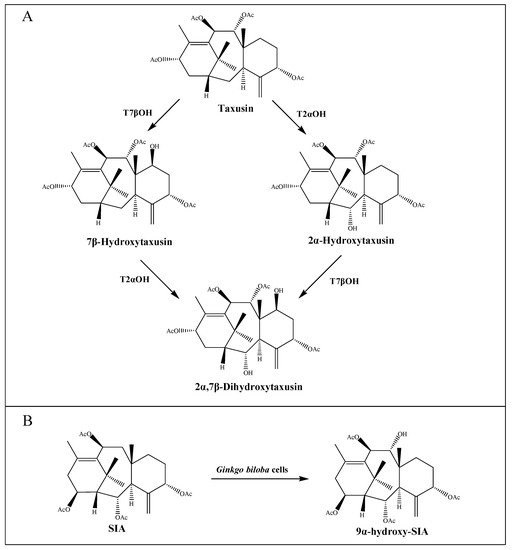
Figure 2. Putative intermediate steps of the taxol pathway. (A) T2αOH- and T7βOH-mediated hydroxylation reactions. (B) T9αOH-mediated hydroxylation reaction in Ginkgo biloba cells.
Table 1. Characterized key enzymes in the taxol pathway.
| Enzyme Abbreviation | Size (kDa) | Probable Localization | GenBank Number |
|---|---|---|---|
| Geranylgeranyl diphosphate synthase GGPPS | 42 | Plastids | AF081514 |
| Taxadiene synthase TS | 98 | Plastids | AY364469 |
| Taxadiene-5α-hydroxylase T5αOH | 56 | Endoplasmic reticulum | AY289209 |
| Taxane-10β-hydroxylase T10βOH | 56 | Endoplasmic reticulum | AF318211 |
| Taxane-10β-hydroxylase a T10βOH | 55 | Endoplasmic reticulum | AY563635 |
| Taxadiene-13α-hydroxylase T13αOH | 54 | Endoplasmic reticulum | AY056019 |
| Taxane-2α-hydroxylase T2αOH | 55 | Endoplasmic reticulum | AY518383 |
| Taxane-9α-hydroxylase T9αOH | 55 | Endoplasmic reticulum | KF773141 |
| Taxane-7β-hydroxylase T7βOH | 56 | Endoplasmic reticulum | AY307951 |
| Taxadiene-5α-ol-O-acetyl transferase TAT | 49 | Cytosol | AF190130 |
| Taxane-2α-O-benzoyl transferase TBT | 50 | Cytosol | AF297618 |
| 10-deacetylbaccatin III-10-O acetyl transferase DBAT | 49 | Cytosol | AF193765 |
| Baccatin III: 3-amino, 13-phenylpropanoyltransferase BAPT | 50 | Cytosol | AY082804 |
| N-benzoyl transferase DBTNBT | 49 | Cytosol | AF466397 |
| Phenylalanine aminomutase PAM | 76 | Cytosol | AY582743 |
| β-phenylalanyl-CoA ligase b PCL | 59 | Cytosol | KM593667 |
Recently, the differential mechanism of taxol synthesis among different tissues of Taxus mairei [42] and T. cuspidate [24], different cultivars within T. yunnanensis [43][44], and different Taxus species (T. media, T. mairei and T. cuspidata) [45][46] was revealed respectively by transcriptome sequencing technology, which showed huge correlations between the taxol contents and the expression levels of taxol pathway genes. Multiomics analysis further confirmed the regulation of these corresponding enzymes on taxol and its derivatives [47][48][49]. Moreover, different elicitors were used to treat Taxus cells. Among them, exogenous methyl jasmonate (MeJA) elicitation significantly increased the expression levels of several taxol pathway genes, such as the Taxus ACT genes [50][51]. The promoter sequences of related genes contained cis-elements involved in different hormones and abiotic stress responses, which may explain why these factors could stimulate taxane biosynthesis in Taxus species [52]. Additionally, the process was believed to be mainly mediated by a series of specific transcription factors (TF), such as MYB, bHLH, ERF, AP2 and MYC. So far, the divergence roles of two ERF TFs, TcERF12 and TcERF15 act as a repressor and activator of the TS gene [53], the negative regulation of TcMYC2a on genes encoding for TS, TAT, DBTNBT, T5αOH and T13αOH either directly or via ERF regulators depending on JA signaling transduction [54], and the positive regulation of TcWRKY1 on its target gene DBAT [55], have been gradually revealed. Moreover, the related regulatory mechanisms of a phloem-specific TmMYB3 [56], and the comprehensive analysis of the R2R3-MYB TF family [57] have also been successively reported in recent years, which promoted further understanding on taxol biosynthesis. Therefore, the increase of production of taxol could be realized by adjusting the catalytic enzymes at the key sites of the core taxane skeleton, which is also the focus and difficulty in the current synthetic biology of taxol.
References
- Wani, M.C.; Taylor, H.L.; Wall, M.E.; Coggon, P.; McPhail, A.T. Plant antitumor agents. VI. Isolation and structure of taxol, a novel antileukemic and antitumor agent from Taxus brevifolia. J. Am. Chem. Soc. 1971, 93, 2325–2327.
- Wani, M.C.; Horwitz, S.B. Nature as a remarkable chemist: A personal story of the discovery and development of taxol. Anti-Cancer Drug. 2014, 25, 482–487.
- Howat, S.; Park, B.; Oh, I.S.; Jin, Y.; Lee, E.; Loake, G.J. Paclitaxel: Biosynthesis, production and future prospects. New Biotechnol. 2014, 31, 242–245.
- Priyadarshini, K.; Keerthi, A.U. Paclitaxel against cancer: A short review. Med. Chem. 2012, 2, 139–141.
- Holton, R.A.; Somoza, C.; Kim, H.B.; Liang, F.; Biediger, R.J.; Boatman, P.D.; Shindo, M.; Smith, C.C.; Kim, S. First total synthesis of taxol. 1. Functionalization of the B ring. J. Am. Chem. Soc. 1994, 116, 1597–1598.
- Nicolaou, K.C.; Yang, Z.; Liu, J.J.; Ueno, H.; Nantermet, P.G.; Guy, R.K.; Claiborne, C.F.; Renaud, J.; Couladouros, E.A.; Paulvannan, K.; et al. Total synthesis of taxol. Nature 1994, 367, 630–634.
- Danishefsky, S.J.; Masters, J.J.; Young, W.B.; Link, J.T.; Snyder, L.B.; Magee, T.V.; Jung, D.K.; Isaacs, R.C.A.; Bornmann, W.G.; Alaimo, C.A.; et al. Total synthesis of baccatin III and Taxol. J. Am. Chem. Soc. 1996, 118, 2843–2859.
- Fu, Y.; Li, S.; Zu, Y.; Yang, G.; Yang, Z.; Luo, M.; Jiang, S.; Wink, M.; Efferth, T. Medicinal chemistry of paclitaxel and its analogues. Curr. Med. Chem. 2009, 16, 3966–3985.
- Shao, F.; Wilson, I.; Qiu, D. The research progress of taxol in Taxus. Curr. Pharm. Biotechnol. 2021, 22, 360–366.
- Liu, W.C.; Gong, T.; Zhu, P. Advances in exploring alternative Taxol sources. RSC Adv. 2016, 6, 48800–48809.
- Wang, T.; Chen, Y.; Zhuang, W.; Zhang, F.; Shu, X.; Wang, Z.; Yang, Q. Transcriptome sequencing reveals regulatory mechanisms of taxol synthesis in Taxus wallichiana var. mairei. Int. J. Genom. 2019, 2019, 1596895.
- Gond, S.K.; Kharwar, R.N.; White, J.F., Jr. Will fungi be the new source of the blockbuster drug taxol? Fungal Biol. Rev. 2014, 28, 77–84.
- Ajikumar, P.K.; Xiao, W.H.; Tyo, K.E.J.; Wang, Y.; Simeon, F.; Leonard, E.; Mucha, O.; Phon, T.H.; Pfeifer, B.; Stephanopoulos, G. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 2010, 330, 70–74.
- Zhou, K.; Qiao, K.; Edgar, S.; Stephanopoulos, G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat. Biotechnol. 2015, 33, 377–383.
- Besumbes, Ó.; Sauret-Güeto, S.; Phillips, M.A.; Imperial, S.; Rodríguez-Concepción, M.; Boronat, A. Metabolic engineering of isoprenoid biosynthesis in Arabidopsis for the production of taxadiene, the first committed precursor of Taxol. Biotechnol. Bioeng. 2004, 88, 168–175.
- Kovacs, K.; Zhang, L.; Linforth, R.T.; Whittaker, B.; Hayes, C.; Fray, R. Redirection of carotenoid metabolism for the efficient production of taxadiene [taxa-4(5),11(12)-diene] in transgenic tomato fruit. Transgen. Res. 2007, 16, 121–126.
- Anterola, A.; Shanle, E.; Perroud, P.; Quatrano, R. Production of taxa-4(5),11(12)-diene by transgenic Physcomitrella patens. Transgen. Res. 2009, 18, 655–660.
- Li, J.; Mutanda, I.; Wang, K.; Yang, L.; Wang, J.; Wang, Y. Chloroplastic metabolic engineering coupled with isoprenoid pool enhancement for committed taxanes biosynthesis in Nicotiana benthamiana. Nat. Commun. 2019, 10, 1–12.
- Wildung, M.R.; Croteau, R. A cDNA clone for taxadiene synthase, the diterpene cyclase that catalyzes the committed step of taxol biosynthesis. J. Biol. Chem. 1996, 271, 9201–9204.
- Hefner, J.; Ketchum, R.E.; Croteau, R. Cloning and functional expression of a cDNA encoding geranylgeranyl diphosphate synthase from Taxus canadensis and assessment of the role of this prenyltransferase in cells induced for taxol production. Arch. Biochem. Biophys. 1998, 360, 62–74.
- Jarchow-Choy, S.K.; Koppisch, A.T.; Fox, D.T. Synthetic routes to methylerythritol phosphate pathway intermediates and downstream isoprenoids. Curr. Org. Chem. 2014, 18, 1050–1072.
- Roberts, S.C. Production and engineering of terpenoids in plant cell culture. Nat. Chem. Biol. 2007, 3, 387.
- Malik, S.; Cusidó, R.M.; Mirjalili, M.H.; Moyano, E.; Palazón, J.; Bonfill, M. Production of the anticancer drug taxol in Taxus baccata suspension cultures: A review. Process. Biochem. 2011, 46, 23.
- Kuang, X.J.; Sun, S.J.; Wei, J.H.; Li, Y.; Sun, C. Iso-Seq analysis of the Taxus cuspidata transcriptome reveals the complexity of Taxol biosynthesis. BMC Plant Biol. 2019, 19, 210.
- Jennewein, S.; Wildung, M.R.; Chau, M.; Walker, K.; Croteau, R. Random sequencing of an induced Taxus cell cDNA library for identification of clones involved in taxol biosynthesis. Proc. Natl. Acad. Sci. USA 2004, 101, 9149–9154.
- Jennewein, S.; Rithner, C.D.; Williams, R.M.; Croteau, R.B. Taxol biosynthesis: Taxane 13 alpha-hydroxylase is a cytochrome p450-dependent monooxygenase. Proc. Natl. Acad. Sci. USA 2001, 98, 13595–13600.
- Walker, K.; Schoendorf, A.; Croteau, R. Molecular cloning of a taxa-4 (20), 11 (12)-dien-5α-ol-O-acetyl transferase cDNA from Taxus and functional expression in Escherichia coli. Arch. Biochem. Biophys. 2000, 374, 371–380.
- Schoendorf, A.; Rithner, C.D.; Williams, R.M.; Croteau, R.B. Molecular cloning of a cytochrome P450 taxane 10 beta-hydroxylase cDNA from Taxus and functional expression in yeast. Proc. Natl. Acad. Sci. USA 2001, 98, 1501–1506.
- Jennewein, S.; Rithner, C.D.; Williams, R.M.; Croteau, R. Taxoid metabolism: Taxoid 14beta-hydroxylase is a cytochrome P450-dependent monooxygenase. Arch. Biochem. Biophys. 2003, 413, 262.
- Holton, R.A.; Kim, H.B.; Somoza, C.; Liang, F.; Biediger, R.J.; Boatman, P.D.; Shindo, M.; Smith, C.C.; Kim, S.C.; Nadizadeh, H.; et al. First total synthesis of taxol. 2. Completion of the C-ring and D-ring. J. Am. Chem. Soc. 1994, 116, 1599–1600.
- Vongpaseuth, K.; Roberts, S.C. Advancements in the understanding of paclitaxel metabolism in tissue culture. Curr. Pharm. Biotechnol. 2007, 8, 219–236.
- Walker, K.; Croteau, R. Taxol biosynthesis: Molecular cloning of a benzoyl-CoA: Taxane 2α-O-benzoyltransferase cDNA from Taxus and functional expression in Escherichia coli. Proc. Natl. Acad. Sci. USA 2000, 97, 13591–13596.
- Walker, K.; Croteau, R.M. Molecular cloning of a 10-deacetylbaccatin Ⅲ-10-O-acetyl transferase cDNA from Taxus and functional expression in Escherichia coli. Proc. Natl. Acad. Sci. USA 2000, 97, 583–587.
- Walker, K.D.; Klettke, K.; Akiyama, T.; Croteau, R. Cloning, heterologous expression, and characterization of a phenylalanine aminomutase involved in Taxol biosynthesis. J. Biol. Chem. 2004, 279, 53947–53954.
- Ramírez-Estrada, K.; Altabella, T.; Onrubia, M.; Moyano, E.; Notredame, C.; Osuna, L.; VandenBossche, R.; Goossens, A.; Cusido, R.; Palazon, J. Transcript profiling of jasmonate-elicited Taxus cells reveals a β-phenylalanine-CoA ligase. Plant Biotechnol. J. 2016, 14, 85–96.
- Walker, K.; Fujisaki, S.; Long, R.; Croteau, R. Molecular cloning and heterologous expression of the C-13 phenylpropanoid side chain-CoA acyltransferase that functions in taxol biosynthesis. Proc. Natl. Acad. Sci. USA 2002, 99, 12715–12720.
- Walker, K.; Long, R.; Croteau, R. The final acylation step in taxol biosynthesis: Cloning of the taxoid C13-side-chain N-benzoyltransferase from Taxus. Proc. Natl. Acad. Sci. USA 2002, 99, 9166.
- Chau, M.; Croteau, R. Molecular cloning and characterization of a cytochrome P450 taxoid 2alpha-hydroxylase involved in Taxol biosynthesis. Arch. Biochem. Biophys. 2004, 427, 48–57.
- Chau, M.; Jennewein, S.; Walker, K.; Croteau, R. Taxol biosynthesis: Molecular cloning and characterization of a cytochrome P450 taxoid 7 β-hydroxylase. Chem. Biol. 2004, 11, 663–672.
- Zhang, N.; Han, Z.T.; Sun, G.L.; Hoffman, A.; Wilson, I.W.; Yang, Y.F.; Gao, Q.; Wu, J.Q.; Xie, D.; Dai, J.G.; et al. Molecular cloning and characterization of a cytochrome P450 taxoid 9α-hydroxylase in Ginkgo biloba cells. Biochem. Bioph. Res. Commun. 2014, 443, 938–943.
- McElroy, C.; Jennewein, S. Taxol biosynthesis and production: From forests to fermenters. In Biotechnology of Natural Products; Schwab, W., Lange, B., Wüst, M., Eds.; Springer: Cham, Switzerland, 2018; pp. 145–185.
- Hao, D.C.; Ge, G.B.; Xiao, P.G.; Zhang, Y.Y.; Ling, Y.; Hans, E. The first insight into the tissue specific Taxus transcriptome via Illumina second generation sequencing. PLoS ONE 2011, 6, e21220.
- He, C.T.; Li, Z.L.; Zhou, Q.; Shen, C.; Huang, Y.Y.; Mubeen, S.; Yang, J.Z.; Yuan, J.G.; Yang, Z.Y. Transcriptome profiling reveals specific patterns of paclitaxel synthesis in a new Taxus yunnanensis cultivar. Plant Physiol. Biochem. 2018, 122, 10–18.
- Mubeen, S.; Li, Z.L.; Huang, Q.M.; He, C.T.; Yang, Z.Y. Comparative transcriptome analysis revealed the tissue-specific accumulations of taxanes among three experimental lines of Taxus yunnanensis. J. Agric. Food. Chem. 2018, 66, 10410–10420.
- Yu, C.N.; Guo, H.; Zhang, Y.Y.; Song, Y.B.; Pi, E.; Yu, C.L.; Zhang, L.; Dong, M.; Zheng, B.S.; Wang, H.Z.; et al. Identification of potential genes that contributed to the variation in the taxoid contents between two Taxus species (Taxus media and Taxus mairei). Tree Physiol. 2017, 37, 1659–1671.
- Zhou, T.; Luo, X.J.; Yu, C.N.; Zhang, C.C.; Zhang, L.; Song, Y.B.; Dong, M.; Shen, C.J. Transcriptome analyses provide insights into the expression pattern and sequence similarity of several taxol biosynthesis related genes in three Taxus species. BMC Plant Biol. 2019, 19, 33.
- Hao, J.; Guo, H.; Shi, X.A.; Wang, Y.; Wan, Q.H.; Song, Y.B.; Zhang, L.; Dong, M.; Shen, C.J. Comparative proteomic analyses of two Taxus species (Taxus × media and Taxus mairei) reveals variations in the metabolisms associated with paclitaxel and other metabolites. Plant Cell Physiol. 2017, 58, 1878–1890.
- Yu, C.N.; Luo, X.J.; Zhan, X.R.; Hao, J.; Zhang, L.; Song, Y.B.; Shen, C.J.; Dong, M. Comparative metabolomics reveals the metabolic variations between two endangered Taxus species (T. fuana and T. yunnanensis) in the Himalayas. BMC Plant Biol. 2018, 18, 197.
- Yu, C.N.; Zhang, C.C.; Xu, X.Y.; Huang, J.F.; Chen, Y.Y.; Luo, X.J.; Wang, H.Z.; Shen, C.J. Omic analysis of the endangered Taxaceae species Pseudotaxus chienii revealed the differences in taxol biosynthesis pathway between Pseudotaxus and Taxus yunnanensis trees. BMC Plant Biol. 2021, 21, 104.
- Li, S.T.; Zhang, P.; Zhang, M.; Fu, C.H.; Zhao, C.F.; Dong, Y.S.; Guo, A.Y.; Yu, L.J. Transcriptional profile of Taxus chinensis cells in response to methyl jasmonate. BMC Genom. 2012, 13, 295.
- Sun, G.; Yang, Y.; Xie, F.; Wen, J.-F.; Wu, J.; Wilson, I.W.; Liu, H.; Qiu, D. Deep sequencing reveals transcriptome re-programming of Taxus× media cells to the elicitation with methyl jasmonate. PLoS ONE 2013, 8, e62865.
- Jiang, L.Y.; Zhang, K.K.; Lü, X.; Yang, L.Y.; Wang, S.; Chen, D.F.; Yang, Y.F.; Qiu, D.Y. Characterization and expression analysis of genes encoding Taxol biosynthetic enzymes in Taxus spp. J. For. Res. 2021.
- Zhang, M.; Li, S.; Nie, L.; Chen, Q.; Xu, X.; Yu, L.; Fu, C. Two jasmonate-responsive factors, TcERF12 and TcERF15, respectively act as repressor and activator of tasy gene of taxol biosynthesis in Taxus chinensis. Plant Mol. Biol. 2015, 89, 463–473.
- Zhang, M.; Jin, X.; Chen, Y.; Wei, M.; Liao, W.; Zhao, S.; Fu, C.; Yu, L. TcMYC2a, a basic helix–loop–helix transcription factor, transduces JA-signals and regulates taxol biosynthesis in Taxus chinensis. Front. Plant Sci. 2018, 9, 863.
- Li, S.; Zhang, P.; Zhang, M.; Fu, C.; Yu, L. Functional analysis of a WRKY transcription factor involved in transcriptional activation of the DBAT gene in Taxus chinensis. Plant Biol. 2012, 15, 19–26.
- Yu, C.N.; Luo, X.J.; Zhang, C.C.; Xu, X.Y.; Huang, J.F.; Chen, Y.Y.; Feng, S.G.; Zhan, X.R.; Zhang, L.; Yuan, H.W.; et al. Tissue-specific study across the stem of Taxus media identifies a phloem-specific TmMYB3 involved in the transcriptional regulation of paclitaxel biosynthesis. Plant J. 2020, 103, 95–110.
- Hu, X.L.; Zhang, L.S.; Wilson, I.; Shao, F.J.; Qiu, D.Y. The R2R3-MYB transcription factor family in Taxus chinensis: Identification, characterization, expression profiling and posttranscriptional regulation analysis. PeerJ 2020, 8, e8473.
More
Information
Subjects:
Chemistry, Medicinal
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
3.1K
Revisions:
2 times
(View History)
Update Date:
01 Jun 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




