Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Junfeng Ma | + 1861 word(s) | 1861 | 2021-09-22 10:22:58 | | | |
| 2 | Amina Yu | + 1266 word(s) | 3127 | 2021-09-23 03:33:05 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Ma, J. OGT Protein Interaction Network (OGT-PIN). Encyclopedia. Available online: https://encyclopedia.pub/entry/14410 (accessed on 08 February 2026).
Ma J. OGT Protein Interaction Network (OGT-PIN). Encyclopedia. Available at: https://encyclopedia.pub/entry/14410. Accessed February 08, 2026.
Ma, Junfeng. "OGT Protein Interaction Network (OGT-PIN)" Encyclopedia, https://encyclopedia.pub/entry/14410 (accessed February 08, 2026).
Ma, J. (2021, September 22). OGT Protein Interaction Network (OGT-PIN). In Encyclopedia. https://encyclopedia.pub/entry/14410
Ma, Junfeng. "OGT Protein Interaction Network (OGT-PIN)." Encyclopedia. Web. 22 September, 2021.
Copy Citation
Interactions between proteins are essential to any cellular process and constitute the basis for molecular networks that determine the functional state of a cell. With the technical advances in recent years, an astonishingly high number of protein–protein interactions has been revealed. However, the interactome of O-linked N-acetylglucosamine transferase (OGT), the sole enzyme adding the O-linked β-N-acetylglucosamine (O-GlcNAc) onto its target proteins, has been largely undefined. To that end, we collated OGT interaction proteins experimentally identified in the past several decades and created a rigorously curated database OGT-Protein Interaction Network (OGT-PIN).
database
O-GlcNAc
OGT
OGT interactome
protein–protein interactions
1. Introduction
Since its discovery in the early 1980s [1][2], O-linked β-N-acetylglucosamine (O-GlcNAc) has been gradually established as an essential post-translational modification of proteins (i.e., O-GlcNAcylation). Distinct from other types of glycosylation, O-GlcNAc is a unique intracellular monosaccharide modification on serine/threonine residues of nuclear/cytoplasmic and mitochondrial proteins [3][4]. It has been found that O-GlcNAcylation occurs on >5000 proteins spanning a range of species, as can be seen from the rigorously curated database O-GlcNAcAtlas [5]. Numerous evidence has demonstrated that the molecular diversity of O-GlcNAcylated proteins has a fundamental importance in many biological processes in physiology and pathology [6][7][8][9][10][11]. Targeting protein O-GlcNAcylation holds great promise for the development of therapeutic targets and biomarkers [12]. Interestingly, despite the substrate diversity, O-GlcNAcylation is catalyzed by only a pair of enzymes: O-GlcNAc transferase (OGT) adds O-GlcNAc onto proteins while O-GlcNAcase (OGA) removes it from proteins [13][14]. Recent years have witnessed great progress towards the understanding of OGT, with deep structural insights obtained and multiple functions revealed [15][16][17][18][19][20]. However, the modes and mechanisms of how OGT works (e.g., interacting with other proteins) have been intriguing and largely unknown.
Mapping protein–protein interactions (PPIs) is instrumental for understanding both the functions of individual proteins and the functional organization of the cell as a whole [21][22][23]. Given the huge importance, a wide array of methods has been developed to probe PPIs in vitro, ex vivo, or in vivo, including yeast two-hybrid (Y2H), protein microarrays, co-immunoprecipitation, affinity chromatography, tandem affinity purification, fluorescence resonance energy transfers (FRET)-related techniques, X-ray crystallography, NMR spectroscopy, and mass spectrometry-based approaches [24][25][26]. Of note, some high throughput methods, especially those coupled with tandem mass spectrometry (MS/MS) (e.g., affinity purification MS (AP-MS), immunoprecipitation-MS (IP-MS), cross-linking MS (XL-MS), proximity labeling MS (PL-MS), and protein correlation profiling MS (PCP-MS)), have enabled the global characterization of PPIs (i.e., interactomics) [27][28][29]. As a critical protein involved in many biological processes, OGT, together with its binding partners, has been unsurprisingly identified from numerous studies. Of note, such methods have also been specifically tailored for the characterization of OGT-interacting proteins recently [30][31][32][33][34][35].
To accommodate the exponentially increased datasets of PPIs, a plethora of comprehensive and specific databases (e.g., BioGRID [36], APID [37], IntAct [38], HuRI [39], HIPPIE [40], HRPD [41], STRING [42], PlaPPISite [43]) have been constructed. These public repertories categorize hundreds and thousands of PPIs from many species. However, after surveying through the >2000 O-GlcNAc-focused studies published previously [5], we found that only a limited number of OGT-interacting proteins had been described in these databases. Furthermore, information of OGT-interactors is sparsely distributed in multiple repositories, with differential stringency applied. To that end, we compiled a rigorously curated and comprehensive database specifically for interaction proteins of OGT and its orthologues identified (e.g., SXC in Drosophila melanogaster, SEC in plants, and OGT-1 in Caenorhabditis elegans), with the goal to provide researchers a rigorously curated but in-depth database for high-stringency OGT interactors. A webserver OGT-PIN (https://oglcnac.org/ogt-pin/) was also constructed, with the hope to better serve investigators in the glycoscience community and beyond.
2. A Comprehensive and High-Stringency Database of OGT-Interacting Proteins
With the technical advances, a number of comprehensive and specific protein interaction databases have been constructed in recent years. Although a joint International Molecular Exchange (IMEx) consortium curation manual (http://www.imexconsortium.org/curation/; version 1 May 2015) was proposed [44], each database appears to have its focus and covers only a portion of protein entries [45]. Given the quickly evolving techniques (especially those for the high throughput analytical characterization), astronomic amount of data is being produced in an unprecedented manner, rendering construction of a unified database for all protein interactions a daunting task.
In this study, we aimed to create a compendium of interacting proteins of OGT and orthologues identified in the past several decades. The OGT interactome database was built using a combination of data retrieved from public repositories and manual extraction and curation from O-GlcNAc-focused studies (both small-scale and large-scale) published previously (as shown in Figure 1). Specifically, we retrieved OGT-interacting proteins from comprehensive databases and specific ones (including BioGRID, APID, IntAct, HuRI, HIPPIE, STRING, and PlaPPISite) that contain hundreds and/or thousands of protein pairs. Besides OGT, several relatively well-characterized OGT orthologues (including SXC in Drosophila melanogaster, SEC in plants, and OGT-1 in Caenorhabditis elegans) were used for searching. Each database was found to contain only a small portion of OGT interaction proteins (not shown). Moreover, there appeared to be relatively small overlaps between databases. To make a complete compendium, manual extraction and curation (by following the single joint IMEx curation manual) were performed to 2503 O-GlcNAc-centered studies from 1984 to 30 December 2020. The combined dataset gave rise to a total of 2492 experimentally identified interaction proteins (Supplementary Table S1).
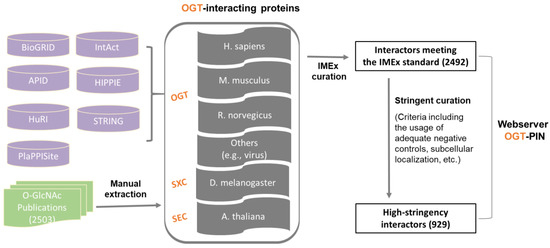
Figure 1. Assembly of the experimentally identified OGT-interacting protein database. In total, information of OGT interactors was extracted from seven public repositories (i.e., BioGRID, APID, IntAct, HuRI, HIPPIE, STRING, and PlaPPISite) and 2503 O-GlcNAc-focused publications. OGT interactors were first curated by following the single joint International Molecular Exchange (IMEx) consortium curation manual and then curated with more stringent criteria (as described in detail in ‘Methods’). Information of both interactors meeting the IMEx standard and interactors with high-stringency was included in the webserver OGT-PIN.
Although the IMEx consortium curation manual provides a basic and general guideline for data curation, we found substantial stringency differences between different datasets. More importantly, there was a need to define high-stringency OGT interactors with more stringent rules. To that end, further curation was performed to the resulting list of proteins meeting the IMEx curation manual. First of all, we noticed that a number of entries from public repertories did not have original sources (e.g., PubMed numbers). Such items were excluded, due to the inability to evaluate those interactors. Moreover, special attention was paid to entries curated from high throughput studies. Although entries from original experiments without adequate negative controls were labeled as ‘Caution’ per the recommendation of IMEx curation manual, such items were excluded from the high-stringency list. In addition, subcellular localization was applied to all entries. Considering OGT interaction has been almost exclusively observed in nuclear/cytosol/mitochondria so far, interactors localized to other organelles (e.g., extracellular space, plasma membrane, Endoplasmic reticulum, and Golgi apparatus) were excluded. This two-step curation strategy yielded a list of 929 high-stringency interaction proteins of OGT and orthologues (from a total of 221 publications) (Supplementary Table S1).
With anticipation, a small overlap between public repertories was observed for the high-stringency OGT interactors (Figure 2). In addition, 620 proteins were additionally curated from O-GlcNAc-centered studies (extracted from 63 publications). One reason for missing so many OGT interaction proteins by public repositories is probably due to the focus of different curators—they may mostly have focused on PPI studies in general while we paid special attention to OGT-involving PPIs from a relatively smaller cohort of publications. These results further demonstrate that there is still a need to construct in-depth protein interaction databases by manual extraction and curation for specific molecules of special interest (as also exemplified for interactomes of phosphatase [46] and heparin/heparan sulfate [47][48]). Last but not least, we believe that data quality of a database is of ultimate importance. Given the false positives reported in the literature (e.g., due to the lack of adequate controls especially in high-throughput proteomic studies), clearly there is a need to treat such entries seriously by curators of every database—to minimize and avoid misleading to users. Adoption of high-stringency and unified curation criteria renders the generation of a database of high confident protein interactors for OGT.
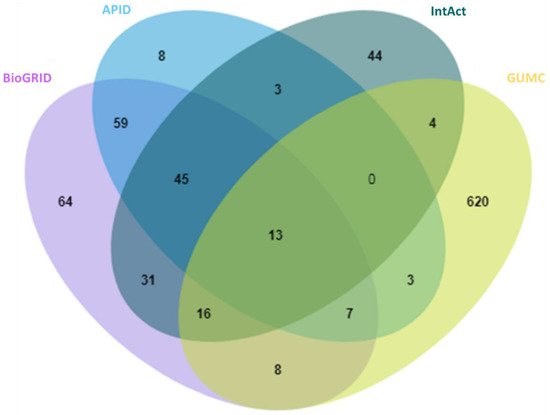
Figure 2. Overlap of 929 high-stringency OGT-interacting proteins from different sources, including BioGRID, APID, IntAct, and GUMC (denoting OGT-interacting proteins additionally extracted and curated by the team at the Georgetown University Medical Center). Curated OGT-interacting proteins obtained from HIPPIE, HuRI, and PlaPPISite were not included for this comparison, due to the presence of only a few additional proteins.
Among all the 929 high-stringency interaction proteins, over half were identified by AP-MS, the most popular tool for protein interatomic studies nowadays. The 929 proteins were found distributed in multiple species investigated, including Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Arabidopsis thaliana, and a few others (e.g., Caenorhabditis elegans, Influenza A virus strains, and SARS-CoV-2). Among them, ~84% (784 human proteins) were interaction partners of human OGT (Figure 3). Moreover, the majority (87%) of OGT interactors were found once, while ~13% were identified by two or more independent studies (Supplementary Figure S1).
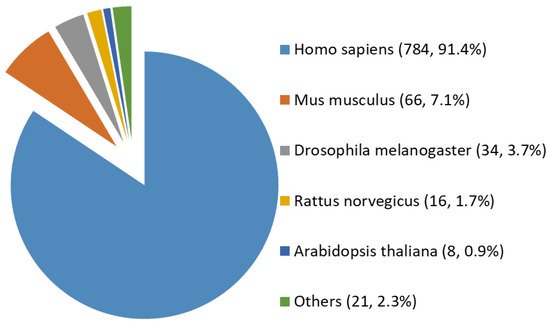
Figure 3. Species distribution of 929 high-stringency interacting proteins of OGT and orthologues. Other interactor proteins include seven from Influenza A virus strains, three from Bacillus anthracis, three from Campylobacter jejuni subsp. jejuni serotype O:2 (strain ATCC 700819/NCTC 11168), two from Caenorhabditis elegans, two from Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) (SARS-CoV-2), one from Human cytomegalovirus (strain Merlin) (HHV-5) (Human herpesvirus 5), one from Oryctolagus cuniculus, one from Francisella tularensis subsp. tularensis (strain SCHU S4/Schu 4), and one from Sus scrofa (pig).
All of the 784 human proteins interacting with human OGT were used to construct a human OGT interactome network. Because the partners of the OGT interactors may interact with each other in vivo, such interactions were extracted by querying STRING. Gephi [49] was used to visualize the OGT network consisting of 782 nodes and 4224 edges (protein interactions) (Figure 4). The highly connected nodes tend to make clusters and hubs in a dense network (the average number of interactions for a node is up to 10.8). A word cloud representation of all interactors of human OGT is shown in Supplementary Figure S2. A list of the top 18 frequently identified interaction proteins is shown in Table 1, including a number of relatively well-characterized interactors by biochemical approaches (e.g., HCFC1, OGT, OGA, TET1, TET2, and TAB1).
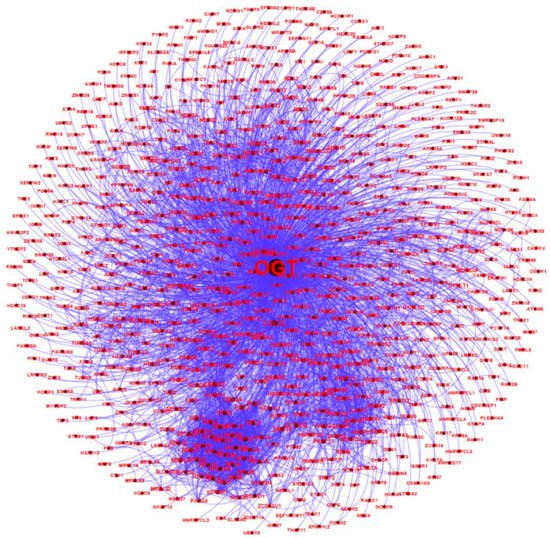
Figure 4. A human OGT interaction network consisting of 782 nodes and 4224 edges (protein interactions). Some nodes are shown in a bigger size due to their higher numbers of associations with other proteins.
Table 1. A list of 18 human OGT-interacting proteins with the highest identification events.
| Entry Name (UniProt) |
Protein Name | Gene Symbol | Number of Times Identified |
|---|---|---|---|
| HCFC1_HUMAN | Host cell factor 1 | HCFC1 | 10 |
| TET2_HUMAN | Methylcytosine dioxygenase TET2 | TET2 | 9 |
| BAP1_HUMAN | Ubiquitin carboxyl-terminal hydrolase BAP1 | BAP1 | 9 |
| OGA_HUMAN | Protein O-GlcNAcase | OGA | 7 |
| SET1A_HUMAN | Histone-lysine N-methyltransferase SETD1A | SETD1A | 6 |
| TRAK1_HUMAN | Trafficking kinesin-binding protein 1 | TRAK1 | 6 |
| OGT1_HUMAN | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | OGT | 6 |
| TET1_HUMAN | Methylcytosine dioxygenase TET1 | TET1 | 6 |
| NUP62_HUMAN | Nuclear pore glycoprotein p62 | NUP62 | 5 |
| WDR5_HUMAN | WD repeat-containing protein 5 | WDR5 | 5 |
| N62CL_HUMAN | Nucleoporin-62 C-terminal-like protein | NUP62CL | 4 |
| SIN3A_HUMAN | Paired amphipathic helix protein Sin3a | SIN3A | 4 |
| DIDO1_HUMAN | Death-inducer obliterator 1 | DIDO1 | 4 |
| RBBP5_HUMAN | Retinoblastoma-binding protein 5 | RBBP5 | 4 |
| TAB1_HUMAN | TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 | TAB1 | 4 |
| TRAK2_HUMAN | Trafficking kinesin-binding protein 2 | TRAK2 | 4 |
| ZEP1_HUMAN | Zinc finger protein 40 | HIVEP1 | 4 |
| TET3_HUMAN | Methylcytosine dioxygenase TET3 | TET3 | 4 |
3. OGT-Interacting Proteins and OGT Substrate Proteins
An intriguing aspect to understand OGT functions is to distinguish its interacting proteins from its substrate proteins. To that end, we compared the 929 high-stringency interaction proteins in our OGT interactome database OGT-PIN with O-GlcNAcAtlas (https://oglcnac.org/; version_01.08), a comprehensive and highly curated database for O-GlcNAc proteins and sites [5]. Very strikingly, it appears that only a small percentage (~39%) of OGT-interacting proteins are OGT substrates (Figure 5), supporting the notion that OGT interactors are not necessarily OGT substrates. Indeed, some of the OGT interactors are also good OGT substrates (e.g., HCFC1, OGT, OGA, TET1, TET2, and TAB1). With the further technical advances in O-GlcNAc site mapping techniques, more OGT-interacting proteins might be found O-GlcNAcylated. But it appears that others (including some highly frequently identified OGT interactors including BAP1, WDR5, FBXW11, and RBBP5 shown in Table 1) are not O-GlcNAcylated. Clearly, the functional roles of such proteins in OGT biology are worthy for further exploration.
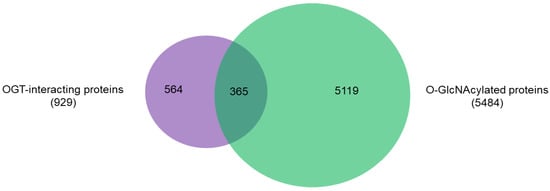
Figure 5. Overlap between 929 high-stringency OGT-interacting proteins in OGT-PIN and O-GlcNAcylated proteins in O-GlcNAcAtlas (Version_01.08).
Recent studies have revealed that OGT is in fact a multi-faceted protein, besides serving as the sole enzyme catalyzing O-GlcNAcylation on thousands of proteins. So far at least four other functions of OGT have been discovered: (1) catalyzes site-specific proteolysis of a transcriptional coactivator HCFC1 [50]; (2) transfers GlcNAc to cysteines (i.e., S-GlcNAc) of cellular proteins [51][52]; (3) use UDP-glucose to install O-linked glucose (O-Glc) onto proteins [53][54]; and (4) catalyzes aspartate to isoaspartate isomerization [55]. The list of high-stringency interaction proteins of OGT will likely provide clues to further understand these non-canonical functions and other functions of OGT yet to be elucidated.
4. Functional Diversity of OGT-Interacting Proteins
The observation that a large proportion (~61%) of interactors are not OGT substrate proteins is very intriguing. Next, we investigated the potential functions of human OGT-interacting proteins. Remarkably, gene ontology (GO) analysis revealed a highly significant enrichment of proteins with the molecular function terms ‘Poly(A) RNA-binding’ and ‘RNA-binding’ (Figure 6A). Concomitantly, ‘transcription’ and ‘(co)translation’ seemed to be the highly enriched biological processes (Figure 6B).
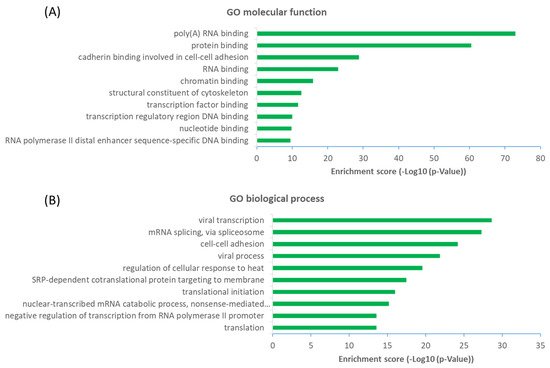
Figure 6. Functional landscape of 784 high-stringency OGT-interacting proteins in human, according to their GO molecular functions (A) and biological processes (B). Only the top ten items with the highest enrichment scores are shown.
From a molecular network perspective, highly clustered modules of OGT interactors were predominantly involved in RNA metabolism, RNA splicing, ribonucleoprotein complexes, chromatin modifications, and others (Figure 7). Since RNA binding proteins play a critical role in controlling various aspects of transcript and translation (including mRNA stability and translation efficiency), the ubiquitous distribution of OGT interactors on transcriptional/translational machinery and other relevant complexes might be a key contributor to the well-documented transcriptional/translational regulation by protein O-GlcNAcylation [10][56][57][58]. Of note, it appears that the interaction partners of OGT were also strongly enriched in proteins involved in cellular responses to stress (Figure 7), in which O-GlcNAcylation has been found to play an important role as well [59].
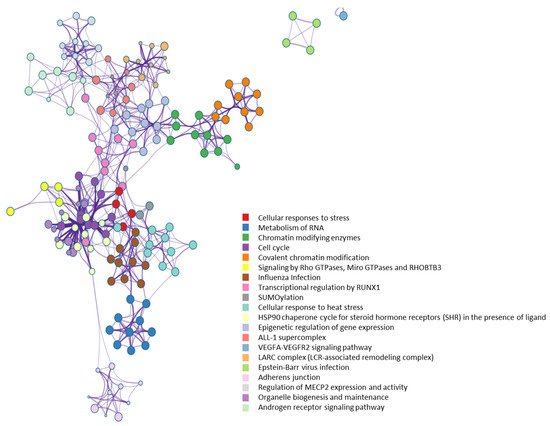
Figure 7. Highly clustered modules of OGT-interacting network. Each term is represented by a circle node, where its size is proportional to the number of input genes that fall into that term, and its color represents its cluster identity (i.e., nodes of the same color belong to the same cluster).
5. OGT as a likely Hub Protein in Cellular Interaction Network
Such a high number of OGT interactors is somewhat unexpected since OGT has not been considered as a hub protein yet [60]. Apparently, OGT has a comparable or even higher number of interacting proteins than many of the ~300 hub proteins (each has several hundred interactors) [60]. Furthermore, OGT is functionally essential since the knockout of OGT is embryonically lethal in a number of organisms [61], fitting well with the classic centrality-lethality rule [62]. Therefore, OGT is likely a hub protein in the cellular network.
The high number of OGT interactors might be closely related to its unique properties. Catalytically, OGT is the sole enzyme that can add O-GlcNAc to thousands of substrate proteins in nuclear, cytosol, and mitochondria. This is distinct from many other enzymes (e.g., glycosyltransferases, kinases, phosphatases, ubiquitin ligases, sirtuins) which often have multiple family members to concertedly modify hundreds or thousands of proteins. It is largely unclear why and how nature chooses OGT to fulfill its duties in such a ubiquitous manner. To achieve that, one possibility is that some OGT interactors serve as scaffold, anchoring, or adaptor proteins that contribute to recruiting active OGT molecules into cellular complexes or by placing OGT close to their substrates, as they do for other post-translational modifications (e.g., phosphorylation) [63][64]. Indeed, among the OGT interactors, quite a few are well-known scaffold proteins (e.g., several 14-3-3 family proteins including YWHAE, YWHAG, YWHAH, and YWHAZ), anchoring proteins (e.g., AKAP2, AKPA12), and adaptor proteins (e.g., importin α). Interestingly, besides binding to OGT, importin α and 14-3-3 proteins also have demonstrated evolutionarily conserved O-GlcNAc binding properties that can directly and selectively recognize/read O-GlcNAc moieties on proteins [65][66][67].
A structural perspective may help partially explain why OGT has so many interactors. OGT has mainly two regions: An N-terminal region consisting of a series of tetratricopeptide repeat (TPR) units (containing 34 amino acids in each) and a multi-domain catalytic C-terminal region. The TPR domains of proteins generally mediate protein–protein interactions and the assembly of multiprotein complexes [68][69]. Although the TPR structural motif is present in many proteins (predicted to be up to 260) [68], human OGT contains a super-helical TPR domain consisting of a very high number of TRP units (13.5). Moreover, the TPR domain appears to be the location where the OGT homotrimer/heterotrimer forms. Crystal structure studies of OGT reveal that TPR superhelix consists of two layers of helices, an inner concave face formed by helix-A and an outer convex face formed by helix-B [15][16][17][18]. The resulting wide binding surface is likely to present several overlapping binding pockets that can hold multiple substrates/interactors. It appears that the conserved asparagine and aspartate ladders regulate the binding of interacting proteins by forming bidentate hydrogen bonds with the peptide backbone [70][71]. In addition, the C-terminal region (e.g., the intervening-D domain and the C-terminal putative phosphatidylinositol-3,4,5-trisphosphate-binding domain) might also be involved in the recognition and binding of versatile proteins [19][20][72].
Despite the great progress especially in the past decade, further studies (e.g., resolving structures of OGT and protein interactors) should promote understanding of detailed interaction mechanisms between OGT and its diverse interacting proteins.
6. Conclusions
The quickly evolving analytical technologies have yielded an enormous amount of protein–protein interaction data, especially in recent years. By combining the datasets from major public repertories and manual extraction and curation of O-GlcNAc-focused studies (both small-scale and large-scale ones), we created a rigorously curated and comprehensive database of OGT-interacting proteins experimentally identified in the past several decades.
Different from public repertories, a two-step curation strategy (by observing both IMEx curation guidelines and our stringent criteria of protein interactors specifically for OGT) was adopted, yielding a list of 929 high-stringency interaction proteins of OGT and orthologues (including 784 proteins interacting with human OGT). Interestingly, only a small percentage (~39%) of OGT-interacting proteins have been identified as OGT substrates. Considering the versatile functions of the diverse interactors, OGT is likely another hub protein in a highly connected cellular network.
We anticipate this reference resource can provide insights into our understanding of OGT biology and protein O-GlcNAcylation. It may also serve as a useful starting point to help with experimental design for further functional elucidation of intracellular proteins/pathways/processes of interest. Given that certain drugs work on the modulation of intracellular protein interaction networks, the resource here may help with translational studies including drug development (e.g., probing the mechanisms of action of drugs and O-GlcNAcylation-targeting therapeutics).
References
- Torres, C.R.; Hart, G.W. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 1984, 259, 3308–3317.
- Holt, G.D.; Hart, G.W. The subcellular distribution of terminal N-acetylglucosamine moieties. Localization of a novel protein-saccharide linkage, O-linked GlcNAc. J. Biol. Chem. 1986, 261, 8049–8057.
- Hart, G.W.; Housley, M.P.; Slawson, C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 2007, 446, 1017–1022.
- Ma, J.; Liu, T.; Wei, A.-C.; Banerjee, P.; O’Rourke, B.; Hart, G.W. O-GlcNAcomic Profiling Identifies Widespread O-Linked β-N-Acetylglucosamine Modification (O-GlcNAcylation) in Oxidative Phosphorylation System Regulating Cardiac Mitochondrial Function. J. Biol. Chem. 2015, 290, 29141–29153.
- Ma, J.; Li, Y.; Hou, C.; Wu, C. O-GlcNAcAtlas: A database of experimentally identified O-GlcNAc sites and proteins. Glycobiology 2021, 31, 719–723.
- Olszewski, N.E.; West, C.M.; Sassi, S.O.; Hartweck, L.M. O-GlcNAc protein modification in plants: Evolution and function. Biochim. Biophys. Acta 2010, 1800, 49–56.
- Hart, G.W.; Slawson, C.; Ramirez-Correa, G.; Lagerlof, O. Cross Talk Between O-GlcNAcylation and Phosphorylation: Roles in Signaling, Transcription, and Chronic Disease. Annu. Rev. Biochem. 2011, 80, 825–858.
- Bond, M.R.; Hanover, J.A. O-GlcNAc cycling: A link between metabolism and chronic disease. Annu. Rev. Nutr. 2013, 33, 205–229.
- Yang, X.; Qian, K. Protein O -GlcNAcylation: Emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2017, 18, 452–465.
- Hart, G.W. Nutrient regulation of signaling and transcription. J. Biol. Chem. 2019, 294, 2211–2231.
- Chatham, J.C.; Zhang, J.; Wende, A.R. Role of O-Linked N-Acetylglucosamine Protein Modification in Cellular (Patho)Physiology. Physiol. Rev. 2020, 101, 427–493.
- Ma, J.; Wu, C.; Hart, G.W. Analytical and Biochemical Perspectives of Protein O-GlcNAcylation. Chem. Rev. 2021, 121, 1513–1581.
- Haltiwanger, R.S.; Holt, G.D.; Hart, G.W. Enzymatic addition of O-GlcNAc to nuclear and cytoplasmic proteins. Identification of a uridine diphospho-N-acetylglucosamine:peptide beta-N-acetylglucosaminyltransferase. J. Biol. Chem. 1990, 265, 2563–2568.
- Dong, D.L.; Hart, G.W. Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. J. Biol Chem. 1994, 269, 19321–19330.
- Jínek, M.; Rehwinkel, J.; Lazarus, B.D.; Izaurralde, E.; Hanover, J.A.; Conti, E. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin α. Nat. Struct. Mol. Biol. 2004, 11, 1001–1007.
- Lazarus, M.B.; Nam, Y.; Jiang, J.; Sliz, P.; Walker, S. Structure of human O -GlcNAc transferase and its complex with a peptide substrate. Nature 2011, 469, 564–567.
- Levine, Z.G.; Walker, S. The Biochemistry of O-GlcNAc Transferase: Which Functions Make It Essential in Mammalian Cells? Annu. Rev. Biochem. 2016, 85, 631–657.
- Rafie, K.; Raimi, O.; Ferenbach, A.T.; Borodkin, V.S.; Kapuria, V.; van Aalten, D.M.F. Recognition of a glycosylation substrate by the O-GlcNAc transferase TPR repeats. Open Biol. 2017, 7.
- King, D.T.; Males, A.; Davies, G.J.; Vocadlo, D.J. Molecular mechanisms regulating O-linked N-acetylglucosamine (O-GlcNAc)–processing enzymes. Curr. Opin. Chem. Biol. 2019, 53, 131–144.
- Joiner, C.M.; Li, H.; Jiang, J.; Walker, S. Structural characterization of the O-GlcNAc cycling enzymes: Insights into substrate recognition and catalytic mechanisms. Curr. Opin. Struct. Biol. 2019, 56, 97–106.
- Barabási, A.-L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113.
- Hu, J.X.; Thomas, C.E.; Brunak, S. Network biology concepts in complex disease comorbidities. Nat. Rev. Genet. 2016, 17, 615–629.
- Caldera, M.; Buphamalai, P.; Müller, F.; Menche, J. Interactome-based approaches to human disease. Curr. Opin. Syst. Biol. 2017, 3, 88–94.
- Fields, S.; Song, O. A novel genetic system to detect protein-protein interactions. Nature 1989, 340, 245–246.
- Rao, V.S.; Srinivas, K.; Sujini, G.N.; Kumar, G.N.S. Protein-Protein Interaction Detection: Methods and Analysis. Int. J. Proteom. 2014, 2014, e147648.
- Xing, S.; Wallmeroth, N.; Berendzen, K.W.; Grefen, C. Techniques for the Analysis of Protein-Protein Interactions in Vivo. Plant Physiol. 2016, 171, 727–758.
- Dunham, W.H.; Mullin, M.; Gingras, A.-C. Affinity-purification coupled to mass spectrometry: Basic principles and strategies. Proteomics 2012, 12, 1576–1590.
- Yu, C.; Huang, L. Cross-Linking Mass Spectrometry: An Emerging Technology for Interactomics and Structural Biology. Anal. Chem. 2018, 90, 144–165.
- Titeca, K.; Lemmens, I.; Tavernier, J.; Eyckerman, S. Discovering cellular protein-protein interactions: Technological strategies and opportunities. Mass Spectrom. Rev. 2019, 38, 79–111.
- Deng, R.-P.; He, X.; Guo, S.-J.; Liu, W.-F.; Tao, Y.; Tao, S.-C. Global identification of O-GlcNAc transferase (OGT) interactors by a human proteome microarray and the construction of an OGT interactome. Proteomics 2014, 14, 1020–1030.
- Ruan, H.-B.; Han, X.; Li, M.-D.; Singh, J.P.; Qian, K.; Azarhoush, S.; Zhao, L.; Bennett, A.M.; Samuel, V.T.; Wu, J.; et al. O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1α stability. Cell Metab. 2012, 16, 226–237.
- Yu, S.-H.; Boyce, M.; Wands, A.M.; Bond, M.R.; Bertozzi, C.R.; Kohler, J.J. Metabolic labeling enables selective photocrosslinking of O-GlcNAc-modified proteins to their binding partners. Proc. Natl. Acad. Sci. USA 2012, 109, 4834–4839.
- Hu, C.-W.; Worth, M.; Xiaofang, Z.; Li, B.; Li, H.; Lu, L.; Zhong, X.; Lin, Z.; Chia-Wei, H.; Ge, Y.; et al. Electrophilic probes for deciphering substrate recognition by O-GlcNAc transferase. Nat. Chem. Biol. 2017, 13, 1267–1273.
- Stephen, H.M.; Praissman, J.L.; Wells, L. Generation of an Interactome for the Tetratricopeptide Repeat Domain of O-GlcNAc Transferase Indicates a Role for the Enzyme in Intellectual Disability. J. Proteome Res. 2021, 20, 1229–1242.
- Martinez, M.; Renuse, S.; Kreimer, S.; O’Meally, R.; Natov, P.; Madugundu, A.K.; Nirujogi, R.S.; Tahir, R.; Cole, R.; Pandey, A.; et al. Quantitative Proteomics Reveals that the OGT Interactome Is Remodeled in Response to Oxidative Stress. Mol. Cell. Proteom. 2021, 20.
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187–200.
- Alonso-López, D.; Campos-Laborie, F.J.; Gutiérrez, M.A.; Lambourne, L.; Calderwood, M.A.; Vidal, M.; De Las Rivas, J. APID database: Redefining protein–protein interaction experimental evidences and binary interactomes. Database 2019, 2019.
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; Del-Toro, N.; et al. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014, 42, D358–D363.
- Luck, K.; Kim, D.-K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Laborie, F.J.C.; Charloteaux, B.; et al. A reference map of the human binary protein interactome. Nature 2020, 580, 402–408.
- Alanis-Lobato, G.; Andrade-Navarro, M.A.; Schaefer, M.H. HIPPIE v2.0: Enhancing meaningfulness and reliability of protein–protein interaction networks. Nucleic Acids Res. 2017, 45, D408–D414.
- Prasad, T.S.K.; Goel, R.; Kandasamy, K.; Keerthikumar, S.; Kumar, S.; Mathivanan, S.; Telikicherla, D.; Raju, R.; Shafreen, B.; Venugopal, A.; et al. Human Protein Reference Database—2009 update. Nucleic Acids Res. 2009, 37, D767–D772.
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612.
- Yang, X.; Yang, S.; Qi, H.; Wang, T.; Li, H.; Zhang, Z. PlaPPISite: A comprehensive resource for plant protein-protein interaction sites. BMC Plant Biol. 2020, 20, 61.
- Orchard, S.; Kerrien, S.; Abbani, S.; Aranda, B.; Bhate, J.; Bidwell, S.; Bridge, A.; Briganti, L.; Brinkman, F.S.L.; Cesareni, G.; et al. Protein interaction data curation: The International Molecular Exchange (IMEx) consortium. Nat. Methods 2012, 9, 345–350.
- Bajpai, A.K.; Davuluri, S.; Tiwary, K.; Narayanan, S.; Oguru, S.; Basavaraju, K.; Dayalan, D.; Thirumurugan, K.; Acharya, K.K. Systematic comparison of the protein-protein interaction databases from a user’s perspective. J. Biomed. Inform. 2020, 103, 103380.
- Sacco, F.; Perfetto, L.; Castagnoli, L.; Cesareni, G. The human phosphatase interactome: An intricate family portrait. FEBS Lett. 2012, 586, 2732–2739.
- Ori, A.; Wilkinson, M.C.; Fernig, D.G. A systems biology approach for the investigation of the heparin/heparan sulfate interactome. J. Biol. Chem. 2011, 286, 19892–19904.
- Gómez Toledo, A.; Sorrentino, J.T.; Sandoval, D.R.; Malmström, J.; Lewis, N.E.; Esko, J.D. A Systems View of the Heparan Sulfate Interactome. J Histochem. Cytochem. 2021, 69, 105–119.
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. Proc. Int. AAAI Conf. Web Soc. Media 2009, 3, 361–362.
- Capotosti, F.; Guernier, S.; Lammers, F.; Waridel, P.; Cai, Y.; Jin, J.; Conaway, J.; Conaway, R.C.; Herr, W. O-GlcNAc Transferase Catalyzes Site-Specific Proteolysis of HCF-1. Cell 2011, 144, 376–388.
- Maynard, J.C.; Burlingame, A.L.; Medzihradszky, K.F. Cysteine S-linked N-acetylglucosamine (S-GlcNAcylation), A New Post-translational Modification in Mammals. Mol. Cell. Proteom. 2016, 15, 3405–3411.
- De Leon, C.A.; Levine, P.M.; Craven, T.W.; Pratt, M.R. The Sulfur-Linked Analogue of O-GlcNAc (S-GlcNAc) Is an Enzymatically Stable and Reasonable Structural Surrogate for O-GlcNAc at the Peptide and Protein Levels. Biochemistry 2017, 56, 3507–3517.
- Shen, D.L.; Liu, T.-W.; Zandberg, W.; Clark, T.; Eskandari, R.; Alteen, M.G.; Tan, H.Y.; Zhu, Y.; Cecioni, S.; Vocadlo, D. Catalytic Promiscuity of O-GlcNAc Transferase Enables Unexpected Metabolic Engineering of Cytoplasmic Proteins with 2-Azido-2-deoxy-glucose. ACS Chem. Biol. 2017, 12, 206–213.
- Darabedian, N.; Gao, J.; Chuh, K.N.; Woo, C.M.; Pratt, M.R. The Metabolic Chemical Reporter 6-Azido-6-deoxy-glucose Further Reveals the Substrate Promiscuity of O-GlcNAc Transferase and Catalyzes the Discovery of Intracellular Protein Modification by O-Glucose. J. Am. Chem. Soc. 2018, 140, 7092–7100.
- Janetzko, J.; Walker, S. Aspartate Glycosylation Triggers Isomerization to Isoaspartate. J. Am. Chem. Soc. 2017, 139, 3332–3335.
- Lewis, B.A.; Hanover, J.A. O-GlcNAc and the epigenetic regulation of gene expression. J. Biol. Chem. 2014, 289, 34440–34448.
- Hardivillé, S.; Hart, G.W. Nutrient Regulation of Signaling, Transcription, and Cell Physiology by O-GlcNAcylation. Cell Metab. 2014, 20, 208–213.
- Parker, M.P.; Peterson, K.R.; Slawson, C. O-GlcNAcylation and O-GlcNAc Cycling Regulate Gene Transcription: Emerging Roles in Cancer. Cancers 2021, 13, 1666.
- Liu, Y.; Yao, R.-Z.; Lian, S.; Liu, P.; Hu, Y.-J.; Shi, H.-Z.; Lv, H.-M.; Yang, Y.-Y.; Xu, B.; Li, S.-Z. O-GlcNAcylation: The “stress and nutrition receptor” in cell stress response. Cell Stress Chaperones 2021, 26, 297–309.
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’Ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128.
- Shafi, R.; Iyer, S.P.N.; Ellies, L.G.; O’Donnell, N.; Marek, K.W.; Chui, D.; Hart, G.W.; Marth, J.D. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. USA 2000, 97, 5735–5739.
- Jeong, H.; Mason, S.P.; Barabási, A.-L.; Oltvai, Z.N. Lethality and centrality in protein networks. Nature 2001, 411, 41–42.
- Pawson, T.; Scott, J.D. Signaling Through Scaffold, Anchoring, and Adaptor Proteins. Science 1997, 278, 2075–2080.
- Zeke, A.; Lukács, M.; Lim, W.A.; Reményi, A. Scaffolds: Interaction platforms for cellular signalling circuits. Trends Cell Biol. 2009, 19, 364–374.
- Seo, H.G.; Kim, H.B.; Kang, M.J.; Ryum, J.H.; Yi, E.C.; Cho, J.W. Identification of the nuclear localisation signal of O -GlcNAc transferase and its nuclear import regulation. Sci. Rep. 2016, 6, 34614.
- Tan, W.; Jiang, P.; Zhang, W.; Hu, Z.; Lin, S.; Chen, L.; Li, Y.; Peng, C.; Li, Z.; Sun, A.; et al. Posttranscriptional regulation of de novo lipogenesis by glucose-induced O-GlcNAcylation. Mol. Cell 2021, 81, 1890–1904.e7.
- Toleman, C.A.; Schumacher, M.A.; Yu, S.-H.; Zeng, W.; Cox, N.J.; Smith, T.J.; Soderblom, E.J.; Wands, A.M.; Kohler, J.J.; Boyce, M. Structural basis of O-GlcNAc recognition by mammalian 14-3-3 proteins. Proc. Natl. Acad. Sci. USA 2018, 115, 5956–5961.
- D’Andrea, L.D.; Regan, L. TPR proteins: The versatile helix. Trends Biochem. Sci. 2003, 28, 655–662.
- Perez-Riba, A.; Itzhaki, L.S. The tetratricopeptide-repeat motif is a versatile platform that enables diverse modes of molecular recognition. Curr. Opin. Struct. Biol. 2019, 54, 43–49.
- Joiner, C.M.; Levine, Z.G.; Aonbangkhen, C.; Woo, C.M.; Walker, S. Aspartate Residues Far from the Active Site Drive O-GlcNAc Transferase Substrate Selection. J. Am. Chem. Soc. 2019, 141, 12974–12978.
- Joiner, C.M.; Hammel, F.A.; Janetzko, J.; Walker, S. Protein Substrates Engage the Lumen of O-GlcNAc Transferase’s Tetratricopeptide Repeat Domain in Different Ways. Biochemistry 2021, 60, 847–853.
- Stephen, H.M.; Adams, T.M.; Wells, L. Regulating the Regulators: Mechanisms of Substrate Selection of the O-GlcNAc Cycling Enzymes OGT and OGA. Glycobiology 2021, 31, 724–733.
More
Information
Subjects:
Cell Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
833
Revisions:
2 times
(View History)
Update Date:
23 Sep 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




