Interactions between proteins are essential to any cellular process and constitute the basis for molecular networks that determine the functional state of a cell. With the technical advances in recent years, an astonishingly high number of protein–protein interactions has been revealed. However, the interactome of O-linked N-acetylglucosamine transferase (OGT), the sole enzyme adding the O-linked β-N-acetylglucosamine (O-GlcNAc) onto its target proteins, has been largely undefined. To that end, we collated OGT interaction proteins experimentally identified in the past several decades and created a rigorously curated database OGT-Protein Interaction Network (OGT-PIN).
1. Introduction
Since its discovery in the early 1980s [
1,
2], O-linked β-N-acetylglucosamine (O-GlcNAc) has been gradually established as an essential post-translational modification of proteins (i.e., O-GlcNAcylation). Distinct from other types of glycosylation, O-GlcNAc is a unique intracellular monosaccharide modification on serine/threonine residues of nuclear/cytoplasmic and mitochondrial proteins [
3,
4]. It has been found that O-GlcNAcylation occurs on >5000 proteins spanning a range of species, as can be seen from the rigorously curated database O-GlcNAcAtlas [
5]. Numerous evidence has demonstrated that the molecular diversity of O-GlcNAcylated proteins has a fundamental importance in many biological processes in physiology and pathology [
6,
7,
8,
9,
10,
11]. Targeting protein O-GlcNAcylation holds great promise for the development of therapeutic targets and biomarkers [
12]. Interestingly, despite the substrate diversity, O-GlcNAcylation is catalyzed by only a pair of enzymes: O-GlcNAc transferase (OGT) adds O-GlcNAc onto proteins while O-GlcNAcase (OGA) removes it from proteins [
13,
14]. Recent years have witnessed great progress towards the understanding of OGT, with deep structural insights obtained and multiple functions revealed [
15,
16,
17,
18,
19,
20]. However, the modes and mechanisms of how OGT works (e.g., interacting with other proteins) have been intriguing and largely unknown.
Mapping protein–protein interactions (PPIs) is instrumental for understanding both the functions of individual proteins and the functional organization of the cell as a whole [
21,
22,
23]. Given the huge importance, a wide array of methods has been developed to probe PPIs in vitro, ex vivo, or in vivo, including yeast two-hybrid (Y2H), protein microarrays, co-immunoprecipitation, affinity chromatography, tandem affinity purification, fluorescence resonance energy transfers (FRET)-related techniques, X-ray crystallography, NMR spectroscopy, and mass spectrometry-based approaches [
24,
25,
26]. Of note, some high throughput methods, especially those coupled with tandem mass spectrometry (MS/MS) (e.g., affinity purification MS (AP-MS), immunoprecipitation-MS (IP-MS), cross-linking MS (XL-MS), proximity labeling MS (PL-MS), and protein correlation profiling MS (PCP-MS)), have enabled the global characterization of PPIs (i.e., interactomics) [
27,
28,
29]. As a critical protein involved in many biological processes, OGT, together with its binding partners, has been unsurprisingly identified from numerous studies. Of note, such methods have also been specifically tailored for the characterization of OGT-interacting proteins recently [
30,
31,
32,
33,
34,
35].
To accommodate the exponentially increased datasets of PPIs, a plethora of comprehensive and specific databases (e.g., BioGRID [
36], APID [
37], IntAct [
38], HuRI [
39], HIPPIE [
40], HRPD [
41], STRING [
42], PlaPPISite [
43]) have been constructed. These public repertories categorize hundreds and thousands of PPIs from many species. However, after surveying through the >2000 O-GlcNAc-focused studies published previously [
5], we found that only a limited number of OGT-interacting proteins had been described in these databases. Furthermore, information of OGT-interactors is sparsely distributed in multiple repositories, with differential stringency applied. To that end, we compiled a rigorously curated and comprehensive database specifically for interaction proteins of OGT and its orthologues identified (e.g., SXC in
Drosophila melanogaster, SEC in plants, and OGT-1 in
Caenorhabditis elegans), with the goal to provide researchers a rigorously curated but in-depth database for high-stringency OGT interactors. A webserver OGT-PIN (
https://oglcnac.org/ogt-pin/) was also constructed, with the hope to better serve investigators in the glycoscience community and beyond.
2. OGT-Interacting Proteins and OGT Substrate Proteins
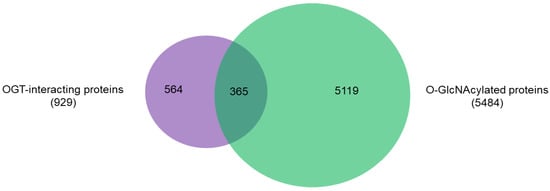
An intriguing aspect to understand OGT functions is to distinguish its interacting proteins from its substrate proteins. To that end, we compared the 929 high-stringency interaction proteins in our OGT interactome database OGT-PIN with O-GlcNAcAtlas (
https://oglcnac.org/; version_01.08), a comprehensive and highly curated database for O-GlcNAc proteins and sites [
5]. Very strikingly, it appears that only a small percentage (~39%) of OGT-interacting proteins are OGT substrates (
Figure 5), supporting the notion that OGT interactors are not necessarily OGT substrates. Indeed, some of the OGT interactors are also good OGT substrates (e.g., HCFC1, OGT, OGA, TET1, TET2, and TAB1). With the further technical advances in O-GlcNAc site mapping techniques, more OGT-interacting proteins might be found O-GlcNAcylated. But it appears that others (including some highly frequently identified OGT interactors including BAP1, WDR5, FBXW11, and RBBP5 shown in
Table 1) are not O-GlcNAcylated. Clearly, the functional roles of such proteins in OGT biology are worthy for further exploration.
Figure 5. Overlap between 929 high-stringency OGT-interacting proteins in OGT-PIN and O-GlcNAcylated proteins in O-GlcNAcAtlas (Version_01.08).
Recent studies have revealed that OGT is in fact a multi-faceted protein, besides serving as the sole enzyme catalyzing O-GlcNAcylation on thousands of proteins. So far at least four other functions of OGT have been discovered: (1) catalyzes site-specific proteolysis of a transcriptional coactivator HCFC1 [
50]; (2) transfers GlcNAc to cysteines (i.e., S-GlcNAc) of cellular proteins [
51,
52]; (3) use UDP-glucose to install O-linked glucose (O-Glc) onto proteins [
53,
54]; and (4) catalyzes aspartate to isoaspartate isomerization [
55]. The list of high-stringency interaction proteins of OGT will likely provide clues to further understand these non-canonical functions and other functions of OGT yet to be elucidated.
3. Functional Diversity of OGT-Interacting Proteins
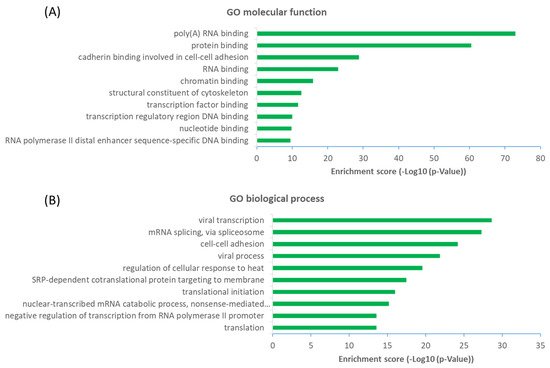
The observation that a large proportion (~61%) of interactors are not OGT substrate proteins is very intriguing. Next, we investigated the potential functions of human OGT-interacting proteins. Remarkably, gene ontology (GO) analysis revealed a highly significant enrichment of proteins with the molecular function terms ‘Poly(A) RNA-binding’ and ‘RNA-binding’ (Figure 6A). Concomitantly, ‘transcription’ and ‘(co)translation’ seemed to be the highly enriched biological processes (Figure 6B).
Figure 6. Functional landscape of 784 high-stringency OGT-interacting proteins in human, according to their GO molecular functions (A) and biological processes (B). Only the top ten items with the highest enrichment scores are shown.
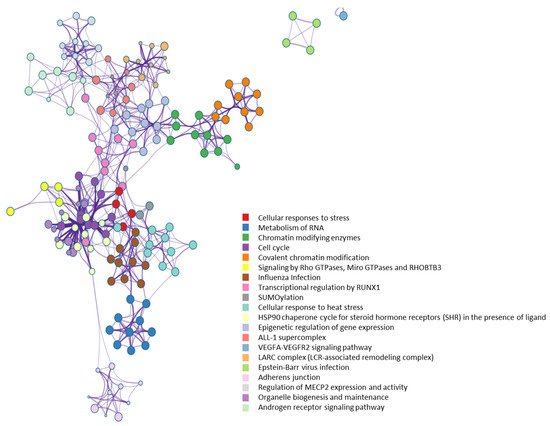
From a molecular network perspective, highly clustered modules of OGT interactors were predominantly involved in RNA metabolism, RNA splicing, ribonucleoprotein complexes, chromatin modifications, and others (
Figure 7). Since RNA binding proteins play a critical role in controlling various aspects of transcript and translation (including mRNA stability and translation efficiency), the ubiquitous distribution of OGT interactors on transcriptional/translational machinery and other relevant complexes might be a key contributor to the well-documented transcriptional/translational regulation by protein O-GlcNAcylation [
10,
56,
57,
58]. Of note, it appears that the interaction partners of OGT were also strongly enriched in proteins involved in cellular responses to stress (
Figure 7), in which O-GlcNAcylation has been found to play an important role as well [
59].
Figure 7. Highly clustered modules of OGT-interacting network. Each term is represented by a circle node, where its size is proportional to the number of input genes that fall into that term, and its color represents its cluster identity (i.e., nodes of the same color belong to the same cluster).
4. OGT as a likely Hub Protein in Cellular Interaction Network
Such a high number of OGT interactors is somewhat unexpected since OGT has not been considered as a hub protein yet [
60]. Apparently, OGT has a comparable or even higher number of interacting proteins than many of the ~300 hub proteins (each has several hundred interactors) [
60]. Furthermore, OGT is functionally essential since the knockout of OGT is embryonically lethal in a number of organisms [
61], fitting well with the classic centrality-lethality rule [
62]. Therefore, OGT is likely a hub protein in the cellular network.
The high number of OGT interactors might be closely related to its unique properties. Catalytically, OGT is the sole enzyme that can add O-GlcNAc to thousands of substrate proteins in nuclear, cytosol, and mitochondria. This is distinct from many other enzymes (e.g., glycosyltransferases, kinases, phosphatases, ubiquitin ligases, sirtuins) which often have multiple family members to concertedly modify hundreds or thousands of proteins. It is largely unclear why and how nature chooses OGT to fulfill its duties in such a ubiquitous manner. To achieve that, one possibility is that some OGT interactors serve as scaffold, anchoring, or adaptor proteins that contribute to recruiting active OGT molecules into cellular complexes or by placing OGT close to their substrates, as they do for other post-translational modifications (e.g., phosphorylation) [
63,
64]. Indeed, among the OGT interactors, quite a few are well-known scaffold proteins (e.g., several 14-3-3 family proteins including YWHAE, YWHAG, YWHAH, and YWHAZ), anchoring proteins (e.g., AKAP2, AKPA12), and adaptor proteins (e.g., importin α). Interestingly, besides binding to OGT, importin α and 14-3-3 proteins also have demonstrated evolutionarily conserved O-GlcNAc binding properties that can directly and selectively recognize/read O-GlcNAc moieties on proteins [
65,
66,
67].
A structural perspective may help partially explain why OGT has so many interactors. OGT has mainly two regions: An N-terminal region consisting of a series of tetratricopeptide repeat (TPR) units (containing 34 amino acids in each) and a multi-domain catalytic C-terminal region. The TPR domains of proteins generally mediate protein–protein interactions and the assembly of multiprotein complexes [
68,
69]. Although the TPR structural motif is present in many proteins (predicted to be up to 260) [
68], human OGT contains a super-helical TPR domain consisting of a very high number of TRP units (13.5). Moreover, the TPR domain appears to be the location where the OGT homotrimer/heterotrimer forms. Crystal structure studies of OGT reveal that TPR superhelix consists of two layers of helices, an inner concave face formed by helix-A and an outer convex face formed by helix-B [
15,
16,
17,
18]. The resulting wide binding surface is likely to present several overlapping binding pockets that can hold multiple substrates/interactors. It appears that the conserved asparagine and aspartate ladders regulate the binding of interacting proteins by forming bidentate hydrogen bonds with the peptide backbone [
70,
71]. In addition, the C-terminal region (e.g., the intervening-D domain and the C-terminal putative phosphatidylinositol-3,4,5-trisphosphate-binding domain) might also be involved in the recognition and binding of versatile proteins [
19,
20,
72].
Despite the great progress especially in the past decade, further studies (e.g., resolving structures of OGT and protein interactors) should promote understanding of detailed interaction mechanisms between OGT and its diverse interacting proteins.
4. Conclusions
The quickly evolving analytical technologies have yielded an enormous amount of protein–protein interaction data, especially in recent years. By combining the datasets from major public repertories and manual extraction and curation of O-GlcNAc-focused studies (both small-scale and large-scale ones), we created a rigorously curated and comprehensive database of OGT-interacting proteins experimentally identified in the past several decades.
Different from public repertories, a two-step curation strategy (by observing both IMEx curation guidelines and our stringent criteria of protein interactors specifically for OGT) was adopted, yielding a list of 929 high-stringency interaction proteins of OGT and orthologues (including 784 proteins interacting with human OGT). Interestingly, only a small percentage (~39%) of OGT-interacting proteins have been identified as OGT substrates. Considering the versatile functions of the diverse interactors, OGT is likely another hub protein in a highly connected cellular network.
We anticipate this reference resource can provide insights into our understanding of OGT biology and protein O-GlcNAcylation. It may also serve as a useful starting point to help with experimental design for further functional elucidation of intracellular proteins/pathways/processes of interest. Given that certain drugs work on the modulation of intracellular protein interaction networks, the resource here may help with translational studies including drug development (e.g., probing the mechanisms of action of drugs and O-GlcNAcylation-targeting therapeutics).
This entry is adapted from the peer-reviewed paper 10.3390/ijms22179620