Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Shunping Yan | + 1245 word(s) | 1245 | 2021-07-01 05:18:08 | | | |
| 2 | Rita Xu | -38 word(s) | 1207 | 2021-07-05 04:58:20 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Yan, S. Chloroplasts. Encyclopedia. Available online: https://encyclopedia.pub/entry/11633 (accessed on 08 February 2026).
Yan S. Chloroplasts. Encyclopedia. Available at: https://encyclopedia.pub/entry/11633. Accessed February 08, 2026.
Yan, Shunping. "Chloroplasts" Encyclopedia, https://encyclopedia.pub/entry/11633 (accessed February 08, 2026).
Yan, S. (2021, July 04). Chloroplasts. In Encyclopedia. https://encyclopedia.pub/entry/11633
Yan, Shunping. "Chloroplasts." Encyclopedia. Web. 04 July, 2021.
Copy Citation
The chloroplast is a semi-autonomous organelle with its own genome. The expression of chloroplast genes depends on both chloroplasts and the nucleus. In chloroplast, NAC102 associates with the chloroplast genome, interacts with chloroplast RNA polymerases, and regulates chloroplast gene expression.
chloroplast
gene expression
NAC102
nucleus
transcription factor
1. Introduction
One of the major differences between plants and animals is that plants contain chloroplasts, which carry out photosynthesis, produce various metabolites, and sense external cues. Chloroplasts are believed to derive from cyanobacterium in an endosymbiotic event. During evolution, most bacteria genes were transferred to the host nucleus and only a small portion of genes were retained on the own genome [1][2]. Therefore, the crosstalk between chloroplasts and the nucleus is essential to maintain cellular homeostasis, which is achieved through anterograde (nucleus-to-chloroplasts) and retrograde (chloroplasts-to-nucleus) signaling [3][4].
As a semi-autonomous organelle, the gene expression in chloroplasts is controlled by both chloroplasts and the nucleus [2]. For example, both plastid-encoded RNA polymerase (PEP) and the nuclear-encoded RNA polymerase (NEP) contribute to chloroplast gene expression [5][6]. While the expression of photosynthetic genes, such as psbA, psbB, and rbcL, is preferentially dependent on PEP, the expression of the housekeeping genes, including ribosomal RNAs and the core subunits of the PEP, is dependent on NEP [7][8][9]. NEP is a phage-type RNA polymerase with a single subunit, encoded by two nuclear genes, rpoTp and rpoTmp, in Arabidopsis. PEP is a bacteria-type RNA polymerase that consists of four core subunits (α, β, β′, and β″), encoded by rpoA, rpoB, rpoC1, and rpoC2 in the chloroplast genome, and a promoter-recognizing sigma factor (σ) encoded by the nuclear genes [10][11]. Furthermore, PEP forms a big complex with many PEP-associated proteins (PAPs) to regulate gene expression. All PAPs are encoded by the nuclear genome [12][13][14].
In addition to PAPs, many other nucleus-encoded proteins have been reported to regulate chloroplast gene expression at the transcriptional level or the post-transcriptional level (including RNA splicing, RNA editing, and translation) [13][15][16]. Although much progress has been made towards understanding their functions in post-transcriptional regulation, how they regulate chloroplast gene expression at the transcriptional level is less well-understood.
In plants, the NAC (NAM, ATAF, and CUC) family is one of the largest plant-specific transcription factors and plays important roles in plant development and stress responses [17][18]. Previous studies have revealed that the transcription factor NAC102 is involved in oxidative stress responses and the nac102 mutant is sensitive to excessive light [19][20][21].
2. Plant Materials and Growth Condition
All Arabidopsis thaliana used in this study were in the Columbia (Col-0) background. The nac102 mutant (SALK_030702) was obtained from ABRC. The transgenic lines were generated by the floral dip method [22]. Arabidopsis was grown on half-strength (1/2) Murashige and Skoog (MS) medium or in soil under long-day conditions (16 h of light and 8 h of dark) at 22 ± 2 °C.
3. Vector Constructions
For localization analysis of NAC102, the coding sequence of enhanced yellow fluorescent protein (eYFP) was fused to the C-terminus of the NAC102 fragment (NAC102-eYFP) using fusion PCR. The NAC102ΔN43-eYFP was generated by using NAC102-eYFP as a template. To generate NAC102N43-eYFP, NAC102N60-eYFP, and NAC102N80-eYFP, the 43, 60 and 80 aa N-terminal parts of NAC102 were cloned and fused to eYFP, respectively. To generate NAC102-eYFPNES, the coding sequence of nuclear export sequence (NES) (CLPPLERLTLD) [23] was further fused C-terminally to NAC102-eYFP (Figure 1). To generate TRXz-CFP, the coding sequence of cyan fluorescent protein (CFP) was fused to the C-terminus of TRXz. All fusion fragments were inserted into pFGC5941 at the NcoI and XbaI sites. For Y2H assays, the coding sequence of NAC102 was inserted into pGBKT7 at the NdeI and PstI sites. The coding sequences of the subunits of PEP and NEP were inserted into pGADT7 at the NdeI and PstI sites. All recombination reactions were performed using a Lightening Cloning kit (Biodragon, Beijing, China).
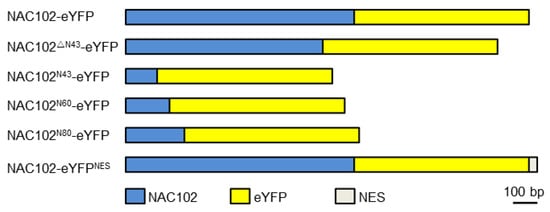
Figure 1. A schematic diagram of NAC102 fusion proteins.
4. NAC102 Localizes in Both Chloroplasts and Nucleus
In Arabidopsis thaliana, there are 105 NAC transcription factors [24], among which NAC102 (AT5G63790) is predicted to localize in both chloroplast and nucleus according to TAIR (https://www.arabidopsis.org; 1 June 2016). To our knowledge, no transcription factors have been reported to localize in the chloroplast. Therefore, we decided to study NAC102 further. NAC102 has two alternative transcripts, AT5G63790.1 and AT5G63790.2. To confirm the subcellular localization of NAC102, both NAC102 variants were fused with enhanced yellow fluorescent protein (eYFP) and were transiently expressed in Arabidopsis protoplasts. The CFP-tagged E2Fa transcription factor (E2Fa-CFP) was used as the nucleus marker and the chlorophyll fluorescence was used as the chloroplast marker. As shown in Figure 2, both AT5G63790.1-eYFP and AT5G63790.2-eYFP localized in chloroplast and nucleus. In the following study, we only used AT5G63790.1. To further confirm its localization, we generated the transgenic Arabidopsis expressing NAC102-eYFP (AT5G63790.1) driven by the 35S promoter (35S:NAC102-eYFP). Consistently, the eYFP fluorescence was detected in both chloroplast and nucleus (Figure 2C).
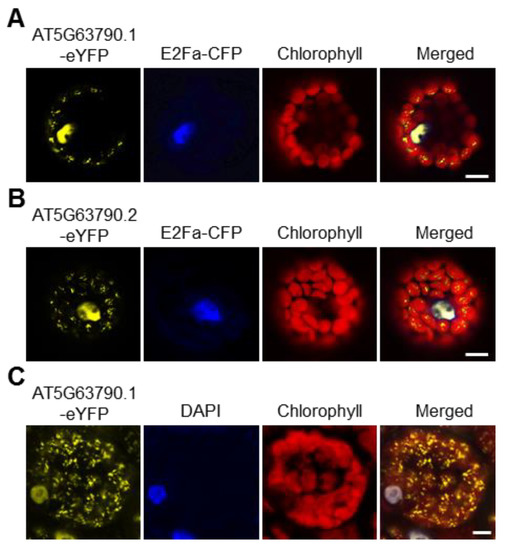
Figure 2. NAC102 localizes in both the nucleus and the chloroplast. (A,B) The localization of NAC102-eYFP when it was transiently expressed in Arabidopsis protoplasts. E2Fa-CFP was used as the nucleus marker and the chlorophyll fluorescence was used as the chloroplast marker. Both isoforms of NAC102, AT5G63790.1 (A) and AT5G63790.2 (B), were shown. (C) The localization of NAC102-eYFP in epidermal cells derived from NAC102 (AT5G63790.1) transgenic lines. The nucleus was stained with DAPI. The pictures were captured using confocal microscopy. Bars = 10 µm.
5. The N-Terminal Sequence of NAC102 Is Necessary and Sufficient for Its Chloroplast Localization
Most nucleus-encoded chloroplast proteins contain transit peptides at their N-terminus to direct them to the chloroplast. To map the transit peptide of NAC102, we performed a prediction using the ChloroP server [25]. However, no transit peptide was identified. Interestingly, when the NAC102 orthologs from other plant species were aligned, we found that the N-terminal 43 amino acid residues (N43) of NAC102 from Arabidopsis thaliana is very unique. We speculated that N43 is necessary for its chloroplast localization. To test this hypothesis, we transiently expressed the truncated NAC102-eYFP without N43 (NAC102ΔN43-eYFP) in Arabidopsis protoplasts. In support of the hypothesis, the NAC102ΔN43-eYFP was only detected in the nucleus (Figure 3A). To investigate whether N43 is sufficient for its chloroplast localization, we fused eYFP with N43 (N43-eYFP) and examined its localization. As shown in Figure 3B, the N43-eYFP could not localize in chloroplasts, suggesting that more amino acid residues are required for the chloroplast localization of NAC102. Therefore, we investigated the localization of eYFP fused to N-terminal 60 or 80 amino acid residues (N60 or N80). We found that the N80-eYFP but not N60-eYFP could localize in chloroplasts (Figure 3C,D). These results suggested that the N-terminal sequence of NAC102 is necessary and sufficient for its chloroplast localization.
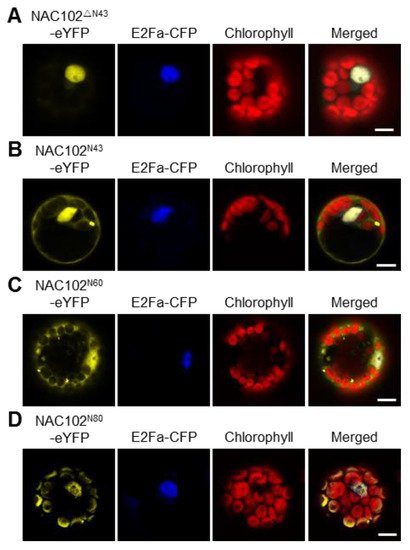
Figure 3. The effects of the N-terminal sequence of NAC102 on its chloroplast localization. The fusion proteins were transiently expressed in Arabidopsis protoplasts. NAC102ΔN43-eYFP (A) represented the truncated NAC102-eYFP without N-terminal 43 amino acid residues (N43); NAC102N43-eYFP (B), NAC102N60-eYFP (C), and NAC102N80-eYFP (D) represented eYFP fused with N-terminal 43, 60 or 80 amino acid residues, respectively. E2Fa-CFP was used as the nucleus marker and the chlorophyll fluorescence was used as the chloroplast marker. The pictures were captured using confocal microscopy. Bars = 10 µm.
References
- Jarvis, P.; Lopez-Juez, E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 2013, 14, 787–802.
- Woodson, J.D.; Chory, J. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 2008, 9, 383–395.
- Chan, K.X.; Phua, S.Y.; Crisp, P.; McQuinn, R.; Pogson, B.J. Learning the languages of the chloroplast: Retrograde signaling and beyond. Annu. Rev. Plant Biol. 2016, 67, 25–53.
- de Souza, A.; Wang, J.Z.; Dehesh, K. Retrograde signals: Integrators of interorganellar communication and orchestrators of plant development. Annu. Rev. Plant Biol. 2017, 68, 85–108.
- Allison, L.A.; Simon, L.D.; Maliga, P. Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J. 1996, 15, 2802–2809.
- Hricova, A.; Quesada, V.; Micol, J.L. The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis. Plant Physiol. 2006, 141, 942–956.
- Hajdukiewicz, P.T.; Allison, L.A.; Maliga, P. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 1997, 16, 4041–4048.
- Liere, K.; Maliga, P. Plastid RNA polymerases in higher plants. Regul. Photosynth. 2011, 11, 29–49.
- Shiina, T.; Tsunoyama, Y.; Nakahira, Y.; Khan, M. Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int. Rev. Cytol. 2005, 244, 1–68.
- Kuhn, K.; Bohne, A.V.; Liere, K.; Weihe, A.; Börner, T. Arabidopsis phage-type RNA polymerases: Accurate in vitro transcription of organellar genes. Plant Cell 2007, 19, 959–971.
- Börner, T.; Aleynikova, A.Y.; Zubo, Y.O.; Kusnetsov, V.V. Chloroplast RNA polymerases: Role in chloroplast biogenesis. Biochim. Biophys. Acta 2015, 1847, 761–769.
- Pfalz, J.; Pfannschmidt, T. Essential nucleoid proteins in early chloroplast development. Trends Plant Sci. 2013, 18, 186–194.
- Yu, Q.B.; Huang, C.; Yang, Z.N. Nuclear-encoded factors associated with the chloroplast transcription machinery of higher plants. Front. Plant Sci. 2014, 5, 316.
- Pfannschmidt, T.; Blanvillain, R.; Merendino, L.; Courtois, F.; Chevalier, F.; Liebers, M.; Grübler, B.; Hommel, E.; Lerbs-Mache, S. Plastid RNA polymerases: Orchestration of enzymes with different evolutionary origins controls chloroplast biogenesis during the plant life cycle. J. Exp. Bot. 2015, 66, 6957–6973.
- Lerbs-Mache, S. Function of plastid sigma factors in higher plants: Regulation of gene expression or just preservation of constitutive transcription? Plant Mol. Biol. 2011, 76, 235–249.
- Zhang, Y.; Zhang, A.; Li, X.; Lu, C. The role of chloroplast gene expression in plant responses to environmental stress. Int. J. Mol. Sci. 2020, 21, 6082.
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87.
- Jensen, M.K.; Skriver, K. NAC transcription factor gene regulatory and protein-protein interaction networks in plant stress responses and senescence. IUBMB Life 2014, 66, 156–166.
- Christianson, J.A.; Wilson, I.W.; Llewellyn, D.J.; Dennis, E.S. The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol. 2009, 149, 1724–1738.
- De Clercq, I.; Van de Velde, J.; Luo, X.; Liu, L.; Storme, V.; Van Bel, M.; Pottie, R.; Vaneechoutte, D.; Van Breusegem, F.; Vandepoele, K. Integrative inference of transcriptional networks in Arabidopsis yields novel ROS signalling regulators. Nat. Plants 2021, 7, 500–513.
- D’Alessandro, S.; Ksas, B.; Havaux, M. Decoding β-Cyclocitral-Mediated Retrograde Signaling Reveals the Role of a Detoxification Response in Plant Tolerance to Photooxidative Stress. Plant Cell 2018, 30, 2495–2511.
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743.
- Heguy, A. Inhibition of the HIV Rev transactivator: A new target for therapeutic intervention. Front Biosci. 1997, 2, 283–297.
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247.
- Emanuelsson, O.; Nielsen, H.; von Heijne, G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999, 8, 978–984.
More
Information
Subjects:
Plant Sciences
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Revisions:
2 times
(View History)
Update Date:
05 Jul 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




