
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Awdhesh Kumar Mishra | + 4122 word(s) | 4122 | 2021-05-10 12:29:08 | | | |
| 2 | Conner Chen | Meta information modification | 4122 | 2021-05-25 09:51:09 | | |
Video Upload Options
Salicylic acid (SA) is an active secondary metabolite that occurs in bacteria, fungi, and plants. SA and its derivatives (collectively called salicylates) are synthesized from chorismate (derived from shikimate pathway). SA is considered an important phytohormone that regulates various aspects of plant growth, environmental stress, and defense responses against pathogens. Besides plants, a large number of bacterial species, such as Pseudomonas, Bacillus, Azospirillum, Salmonella, Achromobacter, Vibrio, Yersinia, and Mycobacteria, have been reported to synthesize salicylates through the NRPS/PKS biosynthetic gene clusters. This bacterial salicylate production is often linked to the biosynthesis of small ferric-ion-chelating molecules, salicyl-derived siderophores (known as catecholate) under iron-limited conditions. Although bacteria possess entirely different biosynthetic pathways from plants, they share one common biosynthetic enzyme, isochorismate synthase, which converts chorismate to isochorismate, a common precursor for synthesizing SA. Additionally, SA in plants and bacteria can undergo several modifications to carry out their specific functions.
1. SA
Phenolic compounds contain an aromatic benzene ring with one or more hydroxyl groups produced as secondary metabolites in nature, primarily in plants and some microorganisms [1][2]. They were presumed to be the byproducts of metabolic pathways, and dispensable for important processes common to all organisms [3]. While plants and a few microorganisms (especially bacteria and fungi) produce phenolics, there are variations between and within species [4]. In plants, phenolics play crucial roles in the regulation of different biochemical and physiological processes [5]. One such important phenolic compound is 2-hydroxy benzoic acid, called salicylic acid (SA) [6][7]. There is a wide variation in the basal levels of SA, with up to 100-fold differences among plants of the same family, and significant differences between species [8][9].
Although SA or its related metabolites have long been used as a pain reliever, the active extract of the inner bark of the Salix alba was isolated and named ‘salicin’ by a German chemist, Johann Buchner, in 1828 [10]. However, the first report of SA signaling in plants was published in 1987, when a mass spectroscopic analysis of the male flowers of calla lily indicated their role in heat production [11]. Subsequent studies revealed that the reproductive organ of gymnosperms and angiosperms displays a thermogenesis phenomenon due to SA signaling [8][12]. A few years after discovering its role in thermogenesis, SA has emerged as a signaling molecule during pathogen infection [13]. Exogenous SA application into tobacco plant leaves was shown to induce pathogenesis-related proteins and improve resistance to tobacco mosaic virus (TMV) infections [14][15]. Since then, many research groups have demonstrated that the increased levels of SA are associated with the induction of defense genes and systemic acquired resistance in the plant species [16][17]. Afterwards, many studies have established that SA is a key signal molecule in regulating the activation of local and systemic defense responses against infections by pathogens [12][16][18]. With SA being accepted as the ‘sixth’ principal plant hormone in the late 1990s [19], several scientific groups started working on its different physiological roles, barring thermogenesis and plant immunity [16][20][21][22][23]. In a short period of time, SA has become an essential signaling molecule in plants and plays a regulatory role in abiotic stresses, like heat stress and drought, and biotic stresses, such as the systemic acquired resistance mediated defense response against pathogen infection [24][25][26][27][28][29]. In addition, SA’s function also influences plant growth and development by regulating various processes, such as photosynthesis, respiration, vegetative growth, seed germination, flowering, senescence, etc. [30][31][32][33]. Furthermore, acetylsalicylic acid (under the popular trade name, Aspirin) has been an important agent for treating various medical conditions [34]. It has been widely used to manage fever and pain, as well as for the management of cardiovascular diseases [34][35][36] and dermatological conditions such as acne, blisters and pruritus [37].
Chorismate (the end-product of the shikimate pathway) is employed as a primary source for the SA biosynthetic pathway. The shikimate pathway starts with erythrose-4-phosphate and phosphoenolpyruvate, and a series of condensation/redox reactions occur, resulting in the formation of chorismate [38][39]. This exclusively conserved pathway is found in bacteria, plants, and fungi, but not in animals [40]. Chorismate is a central metabolic route for the biosynthesis of aromatic amino acids (L-tryptophan, L-phenylalanine, and L-tyrosine) and various aromatic secondary metabolites (such as alkaloids, flavonoids, lignins, aromatic antibiotics, and SA) [41][42]. Thus, chorismate acts as a common connecting precursor of primary and secondary metabolism [43][44]. The first step in the SA biosynthesis is to convert chorismate to isochorismate by using isochorismate synthase (ICS), or its homologous enzyme, common to both bacteria and plants [20][45]. Seven chorismate-utilizing enzymes exist. Of these, five (ICS, salicylate synthase (SAS), anthranilate synthase, aminodeoxyisochorismate synthase, aminodeoxychorismate synthase) are structural homologues and are collectively known as menaquinone, siderophore, and tryptophan (MST) enzymes [46]. The others, two chorismate-utilizing enzymes (chorismate mutase and chorismate lyase), belong to the non-MST family and are responsible for biosynthesizing phenylalanine, tyrosine amino acid, and ubiquinone. Among the above-mentioned seven enzymes, five are present in both plants and bacteria, while the other two (SAS and chorismate lyase) are exclusively reported in bacteria.
The biosynthesis of SA and its functions in plants have now been completely understood through two breakthrough studies [47][48]. However, knowledge on microbial SA biosynthesis is still scarce, and their functions are yet to be elucidated. In plants, SA functions as a hormone, regulating several physiological processes, such as biotic and abiotic stress responses, seed germination, and flowering [6][24][25]. Meanwhile, in bacteria, SA is mainly associated with salicyl-derived siderophores [49][50], and some of its derivative compounds also act as antibacterial agents, e.g., promysalin [51]. Salicyl-derived siderophores are produced by few plant-growth-promoting rhizobacteria and pathogenic bacteria [52][53]. Considering the practical importance of SA, the objective of this study is to investigate the occurrence of bacterial salicylate, its biosynthetic pathways, and compare with plant SA biosynthesis.
2. Biosynthesis of SA: Overview
SA is usually known as a defense-related plant hormone, regulating various cellular processes. In addition, many bacterial spp. also produce SA as an intermediate compound and they are ultimately incorporated into salicylate-based siderophores. All prokaryotic and eukaryotic organisms require chorismate as a starting precursor for SA biosynthesis. In plants, SA biosynthesis is considered to originate from two pathways: the isochorismate synthase (ICS) pathway (major fraction) and the phenylalanine ammonia-lyase (PAL) pathway [6][47][48]. Both are biosynthetic pathways starting in plastids from chorismate and vary between plant species. In bacteria and fungi, SA or its derivates are produced as natural products [49]. These natural products are secondary metabolites (such as SA, siderophore, aromatic antibiotics, and lignins) and encoded by a set of genes termed as a biosynthetic gene cluster (BGC) [54]. Typically, BGC is found as contiguous clusters on the genome, and their genes encode all enzymes required to synthesize a secondary metabolite. All genes are organized as an operon and expressed jointly [55][56]. There are two common BGC systems: nonribosomal peptide synthetase (NRPS) and polyketide synthases (PKS), which are involved in the majority of secondary metabolite synthesis [57]. The borders of a BGC can be hard to predict because multiple genes of a biosynthetic pathway are often expressed on a single operon and separated by few (30–50) nucleotides. In some scenarios, the genes of a BGC are encoded by multiple operons. However, many computational tools, such as ClusterFinder, antiSMASH, ClustScan, etc., can be used to detect BGCs in the microbial genome [58].
2.1. Biosynthesis of SA in Plants
Two SA biosynthetic pathways have been elucidated in plants, namely, the ICS and the PAL pathways [6][20][47][48] (Figure 1). Although plants use both biosynthetic pathways simultaneously, ICS is the major pathway contributing to more than 90% of SA synthesis [48][59]. Both pathways originate from a primary metabolic precursor, chorismate. In the ICS pathway, the first step is the conversion of chorismate to its isomer, isochorismate, by the ICS enzyme, and this is found to be a common step in both bacteria and plants [60]. Usually, plants have one or two genes encoding for the ICS enzyme (e.g., Arabidopsis and soybean have two ICS genes, while rice contains only a single ICS gene). Although their activity may vary among plant species, their primary structures are almost conserved [60]. In plants, isochorismate is synthesized in plastids and subsequently transported from plastids to the cytosol with the enhanced disease susceptibility 5 (EDS5) gene. This gene localized on the plastid envelope and encoded for plastidal MATE (multidrug and toxic compound extrusion) transporter enzyme [47][48][61]. Subsequently, ICS is conjugated with L-glutamate and converted to isochorismate-9-glutamate (ICS-Glu) through the cytosolic amidotransferase enzyme. These amidotransferases are encoded by avrPphB susceptible 3 (PBS3; also known as Gretchen Hagen 3.12). Finally, the spontaneous decomposition of ICS-Glu yields SA and 2-hydroxy- acryloyl-N-Glutamate. However, occasionally, an acyltransferase encoded by enhanced Pseudomonas susceptibility 1 (EPS1) may be involved in this final step [22][48]. Moreover, the activity of PBS3 amidotransferase is inhibited by SA as a negative feedback regulation. Additionally, plants utilize the PAL pathway to synthesize a minor fraction (~10%) of SA and this occurs entirely in the cytosol. Here, the PAL enzyme converts phenylalanine to trans-cinnamic acid (t-CA), and the gene encoding for this enzyme is present in multiple copies in plants [62]. Later, t-CA is converted to SA via two possible intermediates: ortho-coumaric acid and benzaldehyde.
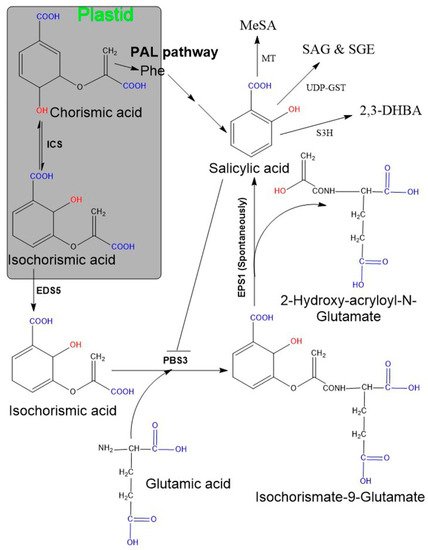
Figure 1. Salicylic acid biosynthesis in plants. Plants possess two pathways (ICS and PAL) and both start from chorisimic acid. In the ICS pathway isochorismate requires two additional genes, named EDS5 and PSB3. The EDS5-encoding enzyme exports isochorismate from the plastid to the cytosol. Further, isochorismate attached to glutamic acid with the help of PBS3 (encoding cytosolic amidotransferase), consequently leads to the formation of an unstable compound, isochorismate-9-glutamate. The non-enzymatic decomposition of isochorismate-9-glutamate then yields salicylate and 2-hydroxy-acryloyl-N-glutamate as final products. The PAL pathway includes the amino acid, phenylalanine as an intermediate compound and comprises multiple sequential enzymatic steps, indicated by several arrows. SA may exist in various modified functional forms, for instance, SAG/SGE, MeSA, 2,3-DHBA. Abbreviation: EDS5 (enhanced disease susceptibility 5), EPS1 (enhanced Pseudomonas susceptibility 1), PBS3 (avrPphB susceptible 3), SAG (SA glucoside), SGE (SA glucose ester), MeSA (methyl salicylate), 2,3-DHBA (2,3-dihydroxybenzoic acid), MT (methyltransferases), UDP-GST (uridine diphosphate glucose-glycosyltransferase) S3H (SA 3-hydroxylase).
SA in plants exists in two main forms: its active free form and its inactive vacuolar storage form (SA glucoside: (SAG), SA glucose ester: (SGE)). SAG and SGE accumulate in the cell vacuoles in large quantities and can form active, usable forms by hydrolysis [63]. Due to pathogen attack, the total SA (SA + SAG/SGE) level increases enormously and activates the systemic acquired resistance dependent defense pathway. In addition, methylation of SA results in the formation of a volatile form of SA, namely methyl salicylate, which is responsible for increased membrane permeability. Methyl salicylate can serve an important function for plant–insect interactions and systemic acquired resistance signaling [64]. Additionally, hydroxylation of SA results in 2,3-dihydroxybenzoic acid (2,3-DHBA) [65].
2.2. Biosynthesis of SA in Bacteria
A large number of different bacterial genera have been reported to synthesize SA, or its related metabolites, especially in plant-growth-promoting rhizobacteria [52]. Among these, the Pseudomonas genus is versatile and its presence is well documented. Bacterial SA synthesis seems to be an artifact and usually incorporates SA into SA-derived siderophores (detail discussed in Section 3). In bacteria, secondary metabolite biosynthesis is controlled by a set of two or more locally clustered genes, known as BGCs. They together encode a core biosynthetic enzyme for the backbone of the compound [66]. In addition, BGC also consists of many genes for (1) regulatory enzymes (transcription factors), (2) tailoring enzymes for modifying backbone structure, and (3) enzymes required for transportation and resistance [67]. Diverse structural classes of BGCs exist, such as nonribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs), bacteriocins, cyclopeptides, terpenes and homoserine lactones, etc. Typically, one BGC accounts for the production of one or several similar bioactive compounds, and they may vary in terms of specificity. Furthermore, the synthesis of some secondary metabolites implicates two or more BGCs.
3. SA-Derived (Catecholate) Siderophores in Bacteria
Siderophores are small Fe3+-chelating secondary metabolites secreted by bacteria and fungi under low-iron conditions [68]. In addition, some plants also produce siderophores for iron acquisition [69]. Generally, siderophores are produced intracellularly and secreted outside the cell as iron-free (deferri) compounds. After scavenging iron (Fe3+ form), the iron-siderophore complex is transported into the cell by the transport system. Based on their structural features and iron-chelating functional groups, siderophores have been classified into three main classes: catecholate (also termed as pheno-catecholate or salicyl-derived siderophore), hydroxamate, and carboxylate [70]. In addition, there are mixed-types (containing more than one of the above-named moieties). In the aforementioned three major classes of siderophores, catecholate siderophores are exclusively produced by bacteria. Usually, bacterial SA is assimilated into the salicyl-derived (termed as salicylates) siderophore backbone. A few mycorrhizal fungi (Ustilago maydis and Phialocephala fortinii) have also been reported to produce hydroxamate siderophore [71]. Further, the Pseudomonas spp. have been documented as the main synthesizer for the catecholate siderophore. More than 100 salicylate siderophores have been reported, and a few are listed in Table 1. Based on their major structural moieties, the catecholate siderophores can be classified into three classes, oxazoline/oxazole, thiazoline/thiazole, and serine-backbone groups. All catecholate siderophores use SA or its hydroxylated derivate, 2,3-dihydroxybenzoic acid (2,3-DHBA), as the common precursor, and its biosynthesis has been well established to involve enzymatic transformations starting from chorismate [72]. Furthermore, several bacteria are capable of producing more than one type of siderophore, such as mycobacterium (produces both types, catecholate and carboxylate).
Table 1. Salicylic acid-producing bacteria and their salicylate-derived siderophores.
| Bacteria Species | Salicylate-Siderophore | Bacterial Source | NRPS Biosynthetic Genes | References |
|---|---|---|---|---|
| Anabaena cylindrical # | Anachelin | Pond | ICS-IPL | [73][74] |
| Acinetobacter baumannii | Acinetobactin * | Human | ICS-IPL | [75][76] |
| Pseudomonas fluorescens CHA0 | Pyochelin | Tobacco rhizosphere | ICS-IPL | [77][78][79] |
| P. fluorescens WCS374 | Pyochelin | Potato rhizosphere | ICS-IPL | [80][81] |
| P. fluorescens WCS417 | Pyochelin | Wheat rhizosphere | ICS-IPL | [80][82][83] |
| Serratia marcescens | Pyochelin | Cucumber/Tobacco rhizosphere | ICS-IPL | [84][85] |
| P. aeruginosa 7NSK2 | Pyochelin | Barley roots | ICS-IPL | [78][86][87][88] |
| P. aureofaciens 63-28 | Pyochelin | Cucumber roots | ICS-IPL | [89] |
| P. corrugata 13 | Pyochelin | Cucumber roots | ICS-IPL | [89] |
| P. fluorescens Pf4–92 | Pseudobactin * | Chickpea rhizosphere | ICS-IPL | [90][91] |
| P. fluorescens PICF3 | Pyochelin | Olive root | ICS-IPL | [81] |
| P. aeruginosa RsG18 and P. aeruginosa RsG27 | Pseudobactin * | Rhizosphere soil | ICS-IPL | [92] |
| P. cepacia | Azurechelin * | Human | ICS-IPL | [93] |
| P.tremae | N/A | Leaves of Salix babylonica | N/A | [94] |
| Paenibacillus larvae | Bacillibactin | Larvae of honeybees | ICS-IPL | [95] |
| Azospirillum iipoferurn D-2 | N/A | Digireria roots | ICS-IPL | [96] |
| Bacillus anthracis | Petrobactin | Sunflower soil | ICS-IPL | [97] |
| Marinobacter hydrocarbonoclasticus | Petrobactin | N/A | ICS-IPL | [98] |
| B. pumilus SF3 | Bacillibactin | Sunflower plant | ICS-IPL | [95][99][100] |
| B. subtilis | Bacillibactin | Banana plant | [100] | |
| Citrobacter | Enterobactin | Tomato roots | ICS-IPL | [101] |
| Klebsiella pneumoniae | Enterobactin | Tomato leaves | ICS-IPL | [101] |
| Photorhabdus luminescens | Photobactin | Nematode(Heterorhabditis bacteriophora) | SAS | [102] |
| Amycolatopsis methanolica 239T | Amychelin | Soil | SAS | [103] |
| Salmonella enterica serotype Typhimurium | Salmochelin | Human | SAS | [104][105][106] |
| Vibrio cholerae | Vibriobactin | Human | SAS | [107][108] |
| V. vulnificus | Vulnibactin | Human | SAS | [108][109][110][111] |
| Yersinia enterocolitica | Yersiniabactin * | Human | SAS | [112][113][114] |
| Y. pestis | Yersiniabactin * | Human | SAS | [115] |
| Mycobacterium tuberculosis | Mycobactin | Human | SAS | [116][117][118][119][120][121] |
* Stands for mixed-type siderophore containing salicylates siderophore; # Stands for cyanobacterium; T Stands for author name (Tang et al.); N/A stands for not available.
Siderophore biosynthesis occurs in two ways: the nonribosomal peptide synthetase and polyketide synthases (NRPS/PKSs) pathway, and the NRPS-independent siderophore (NIS) synthetase pathway. Both NRPS/PKSs and NIS biosynthetic enzymes are encoded as BGCs on the microbial genome. Further, each gene is encoded by a specific module and represents a specific enzyme. Bacteria synthesize salicyl-derived siderophores by two methods: either NRPS biosynthetic gene clusters (NRPS BGC) or NRPS/PKS hybrid biosynthetic gene clusters (NRPS/PKS BGC). For example, pyochelin and bacillibactin biosynthesis involves NRPS, while yersiniabactin and mycobactin involve both NRPS/PKS [122]. PKSs are commonly found in fungi, but they are also contained in a few bacteria. Additionally, some bacteria employ other gene clusters along with NRPS/PKS biosynthetic gene clusters [123].
NRPS catalyzes the synthesis of highly diverse natural microbial products, such as antibiotics, toxins, and siderophores [124]. They are found in all three domains of life (bacteria, archaea, and eukarya) and can synthesize a peptide from a variety of standard, non-proteinogenic amino acids, as well as carboxylic and hydroxy acids. Typically, NRPSs are multimodular enzymes consisting of repeated modules (type I NRPS), but nonmodular enzymes (type II NRPS) are also reported [125][126]. In multimodular NRPS, each module contains three domains: adenylation domain, thiolation domain (also termed as peptidyl carrier protein), and condensation domain. Condensation domains of NRPS (which catalyze the amide bond formation) are functionally the most important and suitable target for natural product genomic analysis [125][127]. Nonmodular NRPSs are linear, comprising specialized tailoring enzymes for more diversification. Nonmodular NRPSs commonly combine their substrate with other pathways to generate a final product [126]. All NRPS biosynthetic genes are organized as operons in BGCs, and their regulation takes place at a transcriptional or post-translational level [128][129]. Additionally, salicylate-derived siderophores comprising BGC show significant similarity among the same bacterial genus. For instance, the salicylate-coding gene cluster of P. fluorescens was compared with the other three Pseudomonas strains and showed a 99% (having 100% query coverage) identity in conserved biosynthetic gene sequences [56].
Biosynthetic genes are encoded by a specific module of the NRPSs gene cluster. Two SA biosynthetic routes have been illustrated in bacteria, and the SA is finally incorporated into salicyl-derived siderophores or other metabolites (Figure 2). First, most SA-producing bacterial spp. (e.g., Pseudomonas and Bacillus spp.) operate an ICS-like biosynthetic pathway. Here, the first step is converting chorismate to its isomer, isochorismate, by the ICS enzyme, while the next step requires another enzyme, isochorismate pyruvate lyase (IPL). IPL converts isochorismate to SA and pyruvate, and is considered a crucial enzyme for bacterial salicylate biosynthesis [45]. So far, no orthologous gene corresponding to bacterial IPL has been identified in plant genomes [7]. Second, some pathogenic bacterial spp. (Yersinia, Vibrio, Salmonella, and Mycobacterium) possess a single bifunctional enzyme called salicylate synthase (SAS) that can directly convert chorismate to SA through an isochorismate intermediate. All bacterial biosynthetic genes, ICS, IPL, and SAS are encoded by a specific module of an NRPS BGC. Further, both ICS and SAS enzymes evolutionally originate from a common ancestor and are highly similar to each other [46][113]. The type I NRPS and the type II NRPS siderophores are represented by the vibriobactin siderophore (in Vibrio) and mycobactin (in Mycobacterium), respectively.
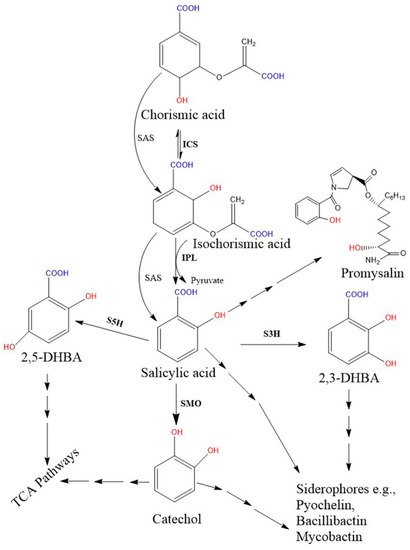
Figure 2. Proposed bacterial pathway for the biosynthesis of SA and their derivative catecholate siderophore. The catecholate siderophore group is only found in bacteria, and their core structures are synthesized from the SA-BGC system. In the Pseudomonas spp., SA is synthesized from chorismate via a two-step enzymatic process (pchA; ICS and pchB; IPL), while the single bifunctional enzyme SAS encoded by MbtI (in Mycobacterium) and Irp9 (in Yersinia) converts chorismate to SA. Further, SMO (also called S1H) converts SA to 1,2-dihydroxybenzene (catechol). Additionally, hydroxylation of SA results in 2,3-DHBA and 2,5 DHBA. Here, salicylate/2, 3-DHBA/catechol can be converted to salicyl-derived siderophores with the help of more than one enzyme. Moreover, catechol and gentisate (2, 5-DHBA) are also metabolized in the TCA pathway. Promysalin, an antibiotic synthesized from SA is also reported in a few Pseudomonas spp. [51]. Solid arrows denote single enzymatic steps, while several arrows denote multiple sequential enzymatic steps. All the demonstrated chemical structures are drawn using the ChemDraw 13.0 software (PerkinElmer, Waltham, MA, USA). Abbreviations: ICS, isochorismate synthase; IPL, isochorismate pyruvate lyase; SAS, salicylate synthase; SMO, salicylate1-monooxygenase; S1H, salicylate 1-hydroxylase; S3H, SA 3-hydroxylase; S5H, salicylate 5-hydroxylase; DHBA, dihydroxybenzoic acid; TCA, tricarboxylic acid.
Salicylate monooxygenase (also known as salicylate 1-hydroxylase) converts salicylate into catechol (1, 2-dihydroxybenzene). This enzyme is also important for salicyl-derived siderophore synthesis and is encoded by a specific module of the NRPS biosynthetic gene clusters [129][130][131]. These enzymes in the Pseudomonas species are encoded from the genes of NahG in P. fluorescens 142 NF and P. putida [131][132], NahW in P. stutzeri AN10 [133], and NahU in P. putida BS3701 [134].
Pyochelin siderophore is made from one molecule of SA and two molecules of cysteine by a thiotemplate mechanism. In Pseudomonas aeruginosa, the biosynthesis of pyochelin takes place by the BGC comprising pchDHIEFKCBA operon [135][136][137]. Two enzymes, pchA (ICS) and pchB (IPL), start the synthesis. Chorismate is converted to isochorismate by the pchA and subsequently, the isochorismate is converted to salicylate by pchB. Subsequently, salicylate is activated by pchD (salicyl-adenylating enzyme). In the meantime, enzyme pchC (encoded for type II thioesterase) removes wrongly charged molecules. Afterward, two tailoring enzymes (pchE and pchF) add L-cysteine residues and perform cyclization and epimerization, resulting in the formation of an intermediate product. Finally, the action of pchK (saccharopine reductase) results in pyochelin synthesis (Figure 3a).
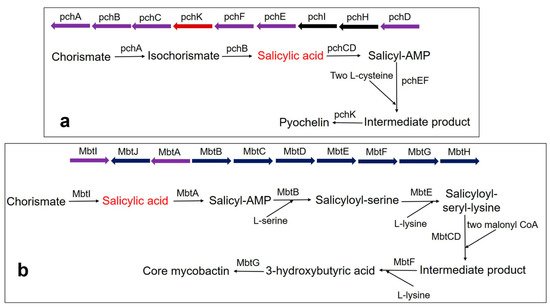
Figure 3. Biosynthesis pathway of catecholate siderophore in bacteria (a) Pyochelin biosynthesis in P. aeruginosa using BGC comprising the pchDHIEFKCBA operon. The biosynthesis of pyochelin is initiated from chorismate by two salicylic acid synthesizing enzymes pchA (ICS) and pchB (IPL), encoded by NRPS BGC. Other gene encoding enzymes are pchC (type II thioesterase); pchD (salicyl-adenylating enzyme); pchE and pchF (tailoring enzyme that adds L-cysteine and performs cyclization and epimerization); pchK (saccharopine reductase); pchH and pchI (transporter enzyme). (b) Core mycobactin scaffold biosynthesis in M. tuberculosis by using BGC comprising the MbtA-J operon. Biosynthesis is carried out by a cluster consisting of 10 essential gene encodings for the enzyme (MbtA-MbtJ). Conversion of chorismate to salicylate requires the main enzyme, MbtI. MbtI (salicylate synthase); MbtA (salicyl-adenylating enzyme); MbtB (enzyme for acyl carrier protein); MbtB and MbtE (enzyme for the addition of serine and lysine residues, respectively); MbtC & MbtD (enzyme for the addition of malonyl CoA residues); MbtF (enzyme for the addition of terminal lysine); MbtG (enzyme for N6-hydroxylation of L-lys); MbtJ and MbtH (enzyme encoding unknown functions).
Mycobactin biosynthesis is performed by an NRPS/PKS hybrid system in M. tuberculosis. The biosynthetic genes are encoded by two BGCs (Mbt-1 and Mbt-2) and include 14 gene encoding enzymes [121]. The Mbt-1 cluster (mbtA-mbtJ) consists of 10 essential gene encodings for biosynthetic enzymes (MbtA-MbtJ) involved in core mycobactin scaffold formation (Figure 3b), while Mbt-2 includes four genes encoding for acyltransferase. Mbt-1 consists of salicylate synthase encoded as Mbt I, which initiates the synthesis of mycobactin followed by the activation of salicylic acid by the adenylating enzyme, MbtA. Similarly, V. cholerae produces the catechol siderophore, vibriobactin, using vibABCDEFH BGC operon. Here, the BGC operon contains SAS encoded by VibH [108][138].
In several bacteria, siderophores can act as virulence factors to combat the host immune system [139][140]. Many catecholate-type siderophores are also found in pathogenic bacteria, such as mycobactin (M. tuberculosis), vibriobactin (Vibrio cholera), salmochelin (Salmonella enterica), petrobactin (Bacillus anthracis), and Yersiniabactin (Yersinia enterocolitica), and are responsible for their pathogenicity (Table 1 [53][141]. Among all the categories of siderophores, the catecholate-type has stronger iron affinities than transferrin and lactoferrin [142]. These features help it to gain a high degree of pathogenicity in catecholate-producing pathogenic bacteria. Pathogens sequester iron from the host protein using siderophores. To counter this, lipocalin-2 protein is released by the host immune system, which sequesters the catecholate-type siderophores and thus impedes bacterial growth [53][143]. Furthermore, few bacteria, such as B. anthracis and S. enterica, secrete lipocalin-2 resistant stealth siderophores and evade the host immune system. Hence, siderophores and their biosynthetic enzymes (ICS/IPL/SAS) could act as a suitable target for drugs and could be helpful for medicine development in the future.
4. Quantification of Salicylic Acid or Salicylate Siderophore
4.1. Quantification of Salicylic Acid in Plant
Quantification of total salicylic acid (both free SA and SA-glucosides) in plant tissues can be done by either using the gas chromatography–mass spectrometry (GC–MS)-based [144][145][146] or the high performance liquid chromatography (HPLC)-based method [147]. For both methods, briefly plant tissues were homogenized in liquid nitrogen and mixed with methanol, followed by centrifugation. Subsequently, the collected supernatant was mixed with extraction solvent such as ethyl acetate–cyclohexane (1:1) (for HPLC-based method) or ethyl acetate (for GC–MS-based method). Afterwards, the sample was analyzed by using a standard amount of SA. GC–MS are highly sensitive approaches that allow more accurate quantification of plant hormones [144].
4.2. Quantification of Bacterial Salicylic Acid and Detection of Salicylate Siderophore
4.2.1. Quantification of Bacterial Salicylic Acid
SA synthesis in broth culture can be determined by the previously described method [78][94]. The chosen bacterial strains were grown in Tris-HCl-buffered (100 mM Tris-HCl; pH 7.5) casamino acid media (0.25 g MgSO4.7H2O, 0.9 g K2HPO4, 5 g casamino acids in one liter) at 37 °C for 24–36 h. Cells were centrifuged at 3500 revs/min for 15 min and the pH of the culture supernatants were adjusted to 2.0 with the help of 1N HCl. Thereafter, SA was extracted by CHCl3 (culture supernatant: CHCl3; 3:1) by vigorous shaking. For the quantitative study, one volume of 2.5 mM FeCl3 was added to the CHCl3 phase. Consequently, the purple Fe–SA complex developed in the aqueous phase, and the absorbance of this complex was measured at 520 nm. SA dissolved in the same growth medium was taken as standard.
4.2.2. Detection of Salicylate Siderophore
Chrome azurol sulphonate (CAS) agar plates were used for regular detection of siderophores [148][149]. However, this assay does not indicate the type of siderophore, and the alternative Arnow’s test was required to be performed for salicylate-derived siderophores [49][150]. This test is based on the reaction between catechol and nitrite-molybdate reagent. The reagent was prepared by dissolving 10 g NaNO2 and 10 g Na2MoO4.2H2O in 100 mL of water. Reactants produce a yellow color in an acidic solution, but their color turns to an intense orange-red in alkaline conditions due to the presence of catecholate-type siderophores. Accordingly, bacterial cultures grown overnight were centrifuged at 3500 revs/min and rinsed with 1X phosphate-buffered saline (pH 7.4). Subsequently, 1.0 mL of culture filtrate was mixed with 1.0 mL of 0.5 N HCl, and 1.0 mL of nitrite-molybdate reagent. Then 1.0 mL of 1N NaOH was added and incubated at room temperature for ~5 min, resulting in the generation of an intense orange-red color formation. The intensity of the color is directly correlated with the amount of siderophores. The absorbance was measured at 510 nm with blank (uninoculated growth media) and standard (known concentration of 2,3-DHBA dissolved in the growth medium).
References
- Cheynier, V. Phenolic compounds: From plants to foods. Phytochem. Rev. 2012, 11, 153–177.
- Van Hung, P. Phenolic compounds of cereals and their antioxidant capacity. Crit. Rev. Food Sci. Nutr. 2016, 56, 25–35.
- Hadacek, F.; Bachmann, G.; Engelmeier, D.; Chobot, V. Hormesis and a chemical raison d’être for secondary plant metabolites. Dose Response 2011, 9, 79–116.
- Mandal, S.M.; Chakraborty, D.; Dey, S. Phenolic acids act as signaling molecules in plant-microbe symbioses. Plant Signal. Behav. 2010, 5, 359–368.
- Wallis, C.M.; Galarneau, E.R.-A. Phenolic Compound induction in plant-microbe and plant-insect interactions: A meta-analysis. Front. Plant Sci. 2020, 11, 2034.
- Lefevere, H.; Bauters, L.; Gheysen, G. Salicylic acid biosynthesis in plants. Front. Plant Sci. 2020, 11, 338.
- Dempsey, D.A.; Vlot, A.C.; Wildermuth, M.C.; Klessig, D.F. Salicylic acid biosynthesis and metabolism. Arab. Book 2011, 9, e0156.
- Raskin, I. Role of salicylic acid in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992, 43, 439–463.
- Seyfferth, C.; Tsuda, K. Salicylic acid signal transduction: The initiation of biosynthesis, perception and transcriptional reprogramming. Front. Plant Sci. 2014, 5, 697.
- Buchner, A. Ueber das rigatellische fiebermittel und über eine in der weidenrinde entdeckte alcaloidische substanz. Repert. Pharm. 1828, 29, 405–420.
- Raskin, I.; Ehmann, A.; Melander, W.R.; Meeuse, B.J.D. Salicylic acid: A natural inducer of heat production in Arum lilies. Science 1987, 237, 1601–1602.
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206.
- Malamy, J.; Carr, J.P.; Klessig, D.F.; Raskin, I. Salicylic acid: A likely endogenous signal in the resistance response of tobacco to viral infection. Science 1990, 250, 1002–1004.
- White, R.F. Acetylsalicylic acid (aspirin) induces resistance to tobacco mosaic virus in tobacco. Virology 1979, 99, 410–412.
- Antoniw, J.F.; White, R.F. The effects of aspirin and polyacrylic acid on soluble leaf proteins and resistance to virus infection in five cultivars of Tobacco. J. Phytopathol. 1980, 98, 331–341.
- Klessig, D.F.; Choi, H.W.; Dempsey, D.A. Systemic acquired resistance and salicylic acid: Past, present, and future. Mol. Plant Microbe Interact. 2018, 31, 871–888.
- Métraux, J.P.; Signer, H.; Ryals, J.; Ward, E.; Wyss-Benz, M.; Gaudin, J.; Raschdorf, K.; Schmid, E.; Blum, W.; Inverardi, B. Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 1990, 250, 1004–1006.
- An, C.; Mou, Z. Salicylic acid and its function in plant immunity. J. Integr. Plant Biol. 2011, 53, 412–428.
- Raskin, I. Salicylate, A New Plant Hormone 1. Plant Physiol. 1992, 99, 799–803.
- Dempsey, D.A.; Klessig, D.F. How does the multifaceted plant hormone salicylic acid combat disease in plants and are similar mechanisms utilized in humans? BMC Biol. 2017, 15, 23.
- Malamy, J.; Klessig, D.F. Salicylic acid and plant disease resistance. Plant J. 1992, 2, 643–654.
- Ding, P.; Ding, Y. Stories of salicylic acid: A plant defense hormone. Trends Plant Sci. 2020, 25, 549–565.
- Gu, X.Y.; Liu, Y.; Liu, L.J. Progress on the biosynthesis and signal transduction of phytohormone salicylic acid. Yi Chuan Hered. 2020, 42, 858–869.
- Zhang, Y.; Li, X. Salicylic acid: Biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 2019, 50, 29–36.
- Khan, M.I.R.; Fatma, M.; Per, T.S.; Anjum, N.A.; Khan, N.A. Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 2015, 6, 462.
- Miura, K.; Tada, Y. Regulation of water, salinity, and cold stress responses by salicylic acid. Front. Plant Sci. 2014, 5, 4.
- Alazem, M.; Lin, N.-S. Roles of plant hormones in the regulation of host–virus interactions. Mol. Plant Pathol. 2015, 16, 529–540.
- Freeman, J.L.; Garcia, D.; Kim, D.; Hopf, A.; Salt, D.E. Constitutively elevated salicylic acid signals glutathione-mediated nickel tolerance in Thlaspi nickel hyperaccumulators. Plant Physiol. 2005, 137, 1082–1091.
- Larkindale, J.; Hall, J.D.; Knight, M.R.; Vierling, E. Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. 2005, 138, 882–897.
- Rivas-San Vicente, M.; Plasencia, J. Salicylic acid beyond defence: Its role in plant growth and development. J. Exp. Bot. 2011, 62, 3321–3338.
- Pasternak, T.; Groot, E.P.; Kazantsev, F.V.; Teale, W.; Omelyanchuk, N.; Kovrizhnykh, V.; Palme, K.; Mironova, V. V Salicylic acid affects root meristem patterning via auxin distribution in a concentration-dependent manner. Plant Physiol. 2019, 180, 1725–1739.
- Lee, S.; Kim, S.; Park, C. Salicylic acid promotes seed germination under high salinity by modulating antioxidant activity in Arabidopsis. New Phytol. 2010, 188, 626–637.
- Van Butselaar, T.; Van den Ackerveken, G. Salicylic acid steers the growth-immunity tradeoff. Trends Plant Sci. 2020, 25, 566–576.
- Weissmann, G. Aspirin. Sci. Am. 1991, 264, 84–91.
- Cuzick, J.; Thorat, M.A.; Bosetti, C.; Brown, P.H.; Burn, J.; Cook, N.R.; Ford, L.G.; Jacobs, E.J.; Jankowski, J.A.; La Vecchia, C.; et al. Estimates of benefits and harms of prophylactic use of aspirin in the general population. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2015, 26, 47–57.
- Dachineni, R.; Kumar, D.R.; Callegari, E.; Kesharwani, S.S.; Sankaranarayanan, R.; Seefeldt, T.; Tummala, H.; Bhat, G.J. Salicylic acid metabolites and derivatives inhibit CDK activity: Novel insights into aspirin’s chemopreventive effects against colorectal cancer. Int. J. Oncol. 2017, 51, 1661–1673.
- Arif, T. Salicylic acid as a peeling agent: A comprehensive review. Clin. Cosmet. Investig. Dermatol. 2015, 8, 455–461.
- Herrmann, K.M. The shikimate pathway: Early steps in the biosynthesis of aromatic compounds. Plant Cell 1995, 7, 907–919.
- Averesch, N.J.H.; Krömer, J.O. Metabolic Engineering of the Shikimate Pathway for Production of Aromatics and Derived Compounds—Present and Future Strain Construction Strategies. Front. Bioeng. Biotechnol. 2018, 6, 32.
- Kayser, N.A.O.; Averesch, N. Technische Biochemie: Die Biochemie und Industrielle Nutzung von Naturstoffen; Springer: Wiesbaden, Germany, 2015; ISBN 3658055480.
- Tzin, V.; Galili, G. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant 2010, 3, 956–972.
- Tohge, T.; Watanabe, M.; Hoefgen, R.; Fernie, A. Shikimate and phenylalanine biosynthesis in the green lineage. Front. Plant Sci. 2013, 4, 62.
- Hubrich, F.; Müller, M.; Andexer, J.N. Chorismate- and isochorismate converting enzymes: Versatile catalysts acting on an important metabolic node. Chem. Commun. 2021, 57, 2441–2463.
- Dosselaere, F.; Vanderleyden, J. A Metabolic Node in Action: Chorismate-utilizing enzymes in microorganisms. Crit. Rev. Microbiol. 2001, 27, 75–131.
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001, 414, 562.
- Shelton, C.L.; Lamb, A.L. Unraveling the structure and mechanism of the MST(ery) enzymes. Trends Biochem. Sci. 2018, 43, 342–357.
- Torrens-Spence, M.P.; Bobokalonova, A.; Carballo, V.; Glinkerman, C.M.; Pluskal, T.; Shen, A.; Weng, J.-K. PBS3 and EPS1 complete salicylic acid biosynthesis from isochorismate in Arabidopsis. Mol. Plant 2019, S1674-2052, 30369–30377.
- Rekhter, D.; Lüdke, D.; Ding, Y.; Feussner, K.; Zienkiewicz, K.; Lipka, V.; Wiermer, M.; Zhang, Y.; Feussner, I. Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 2019, 365, 498–502.
- Grobelak, A.; Hiller, J. Bacterial siderophores promote plant growth: Screening of catechol and hydroxamate siderophores. Int. J. Phytoremediation 2017, 19, 825–833.
- Maruri-López, I.; Aviles-Baltazar, N.Y.; Buchala, A.; Serrano, M. Intra and extracellular journey of the phytohormone salicylic acid. Front. Plant Sci. 2019, 10, 423.
- Kaduskar, R.D.; Della Scala, G.; Al Jabri, Z.J.; Arioli, S.; Musso, L.; Oggioni, M.R.; Dallavalle, S.; Mora, D. Promysalin is a salicylate-containing antimicrobial with a cell-membrane-disrupting mechanism of action on Gram-positive bacteria. Sci. Rep. 2017, 7, 8861.
- Bakker, P.A.H.M.; Ran, L.; Mercado-Blanco, J. Rhizobacterial salicylate production provokes headaches! Plant Soil 2014, 382, 1–16.
- Wilson, B.R.; Bogdan, A.R.; Miyazawa, M.; Hashimoto, K.; Tsuji, Y. Siderophores in iron metabolism: From mechanism to therapy potential. Trends Mol. Med. 2016, 22, 1077–1090.
- Kjærbølling, I.; Mortensen, U.H.; Vesth, T.; Andersen, M.R. Strategies to establish the link between biosynthetic gene clusters and secondary metabolites. Fungal Genet. Biol. 2019, 130, 107–121.
- Tran, P.N.; Yen, M.-R.; Chiang, C.-Y.; Lin, H.-C.; Chen, P.-Y. Detecting and prioritizing biosynthetic gene clusters for bioactive compounds in bacteria and fungi. Appl. Microbiol. Biotechnol. 2019, 103, 3277–3287.
- Shehroz, M.; Aslam, M.; Ali Khan, M.; Aiman, S.; Gul Afridi, S.; Khan, A. The in silico characterization of a salicylic acid analogue coding fene clusters in selected Pseudomonas fluorescens strains. Iran. J. Biotechnol. 2019, 17, e2250.
- Weber, T.; Kim, H.U. The secondary metabolite bioinformatics portal: Computational tools to facilitate synthetic biology of secondary metabolite production. Synth. Syst. Biotechnol. 2016, 1, 69–79.
- Blin, K.; Kim, H.U.; Medema, M.H.; Weber, T. Recent development of antiSMASH and other computational approaches to mine secondary metabolite biosynthetic gene clusters. Brief. Bioinform. 2019, 20, 1103–1113.
- Chen, Z.; Zheng, Z.; Huang, J.; Lai, Z.; Fan, B. Biosynthesis of salicylic acid in plants. Plant Signal. Behav. 2009, 4, 493–496.
- Yokoo, S.; Inoue, S.; Suzuki, N.; Amakawa, N.; Matsui, H.; Nakagami, H.; Takahashi, A.; Arai, R.; Katou, S. Comparative analysis of plant isochorismate synthases reveals structural mechanisms underlying their distinct biochemical properties. Biosci. Rep. 2018, 38, BSR20171457.
- Yamasaki, K.; Motomura, Y.; Yagi, Y.; Nomura, H.; Kikuchi, S.; Nakai, M.; Shiina, T. Chloroplast envelope localization of EDS5, an essential factor for salicylic acid biosynthesis in Arabidopsis thaliana. Plant Signal. Behav. 2013, 8, e23603.
- Dong, C.-J.; Shang, Q.-M. Genome-wide characterization of phenylalanine ammonia-lyase gene family in watermelon (Citrullus lanatus). Planta 2013, 238, 35–49.
- Dean, J.V.; Shah, R.P.; Mohammed, L.A. Formation and vacuolar localization of salicylic acid glucose conjugates in soybean cell suspension cultures. Physiol. Plant. 2003, 118, 328–336.
- Snoeren, T.A.L.; Mumm, R.; Poelman, E.H.; Yang, Y.; Pichersky, E.; Dicke, M. The herbivore-induced plant volatile methyl salicylate negatively affects attraction of the parasitoid Diadegma semiclausum. J. Chem. Ecol. 2010, 36, 479–489.
- Zhang, K.; Halitschke, R.; Yin, C.; Liu, C.-J.; Gan, S.-S. Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc. Natl. Acad. Sci. USA 2013, 110, 14807–14812.
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; De Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 2015, 11, 625–631.
- Medema, M.; Fischbach, M. Computational approaches to natural product discovery. Nat. Chem. Biol. 2015, 11, 639–648.
- Kramer, J.; Özkaya, Ö.; Kümmerli, R. Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 2020, 18, 152–163.
- Kumar, L.; Meena, N.L.; Singh, U. Role of phytosiderophores in acquisition of iron and other micronutrients in food legumes. In Biofortification of Food Crops; Singh, U., Praharaj, C.S., Singh, S.S., Singh, N.P., Eds.; Springer India: New Delhi, India, 2016; pp. 291–302. ISBN 978-81-322-2716-8.
- Khan, A.; Singh, P.; Srivastava, A. Synthesis, nature and utility of universal iron chelator–Siderophore: A review. Microbiol. Res. 2018, 212, 103–111.
- Das, A.; Prasad, R.; Srivastava, A.; Giang, P.H.; Bhatnagar, K.; Varma, A. Fungal Siderophores: Structure, Functions and Regulation BT—Microbial Siderophores; Varma, A., Chincholkar, S.B., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 1–42. ISBN 978-3-540-71160-5.
- Plach, M.G.; Löffler, P.; Merkl, R.; Sterner, R. Conversion of anthranilate synthase into isochorismate synthase: Implications for the Evolution of chorismate-utilizing enzymes. Angew. Chem. Int. Ed. 2015, 54, 11270–11274.
- Beiderbeck, H.; Taraz, K.; Budzikiewicz, H.; Walsby, A.E. Anachelin, the siderophore of the cyanobacterium Anabaena cylindrica CCAP 1403/2A. Z. Nat. C 2000, 55, 681–687.
- Gademann, K.; Kobylinska, J.; Wach, J.-Y.; Woods, T.M. Surface modifications based on the cyanobacterial siderophore anachelin: From structure to functional biomaterials design. BioMetals 2009, 22, 595–604.
- Vallenet, D.; Nordmann, P.; Barbe, V.; Poirel, L.; Mangenot, S.; Bataille, E.; Dossat, C.; Gas, S.; Kreimeyer, A.; Lenoble, P.; et al. Comparative analysis of Acinetobacters: Three genomes for three lifestyles. PLoS ONE 2008, 3, e1805.
- Song, W.Y.; Kim, H.J. Current biochemical understanding regarding the metabolism of acinetobactin, the major siderophore of the human pathogen Acinetobacter baumannii, and outlook for discovery of novel anti-infectious agents based thereon. Nat. Prod. Rep. 2020, 37, 477–487.
- Stutz, E.W.; Défago, G.; Kern, H. Naturally occurring fluorescent pseudomonads involved in suppression of black root rot of tobacco. Phytopathology 1986, 76, 181–185.
- De Meyer, G.; Höfte, M. Salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa 7NSK2 induces resistance to leaf infection by Botrytis cinerea on bean. Phytopathology 1997, 87, 588–593.
- Maurhofer, M. Induction of systemic resistance of tobacco to tobacco necrosis virus by the root-colonizing Pseudomonas fluorescens strain CHA0: Influence of the gacA gene and of pyoverdine production. Phytopathology 1994, 84, 139–146.
- Leeman, M.; Van Pelt, J.A.; Den Ouden, F.M.; Heinsbroek, M.; Bakker, P.A.; Schippers, B. Induction of systemic resistance against Fusarium wilt of radish by lipopolysaccharides of Pseudomonas fluorescens. Phytopathology 1995, 85, 1021–1027.
- Mercado-Blanco, J.; van der Drift, K.M.G.M.; Olsson, P.E.; Thomas-Oates, J.E.; van Loon, L.C.; Bakker, P.A.H.M. Analysis of the pmsCEAB gene cluster involved in biosynthesis of salicylic acid and the siderophore pseudomonine in the biocontrol strain Pseudomonas fluorescens WCS374. J. Bacteriol. 2001, 183, 1909–1920.
- Van Wees, S.C.M.; Pieterse, C.M.J.; Trijssenaar, A.; Van ’t Westende, Y.A.M.; Hartog, F.; Van Loon, L.C. Differential induction of systemic resistance in Arabidopsis by biocontrol bacteria. Mol. Plant Microbe Interact. 1997, 10, 716–724.
- Ran, L.X.; Liu, C.Y.; Wu, G.J.; van Loon, L.C.; Bakker, P.A.H.M. Suppression of bacterial wilt in Eucalyptus urophylla by fluorescent Pseudomonas spp. in China. Biol. Control 2005, 32, 111–120.
- Press, C.M.; Wilson, M.; Tuzun, S.; Kloepper, J.W. Salicylic acid produced by Serratia marcescens 90-166 is not the primary determinant of induced systemic resistance in cucumber or tobacco. Mol. Plant Microbe Interact. 1997, 10, 761–768.
- Khilyas, I.V.; Tursunov, K.A.; Shirshikova, T.V.; Kamaletdinova, L.K.; Matrosova, L.E.; Desai, P.T.; McClelland, M.; Bogomolnaya, L.M. Genome sequence of pigmented siderophore-producing strain Serratia marcescens SM6. Microbiol. Resour. Announc. 2019, 8, e00247-19.
- Buysens, S.; Heungens, K.; Poppe, J.; Hofte, M. Involvement of pyochelin and pyoverdin in suppression of Pythium-induced damping-off of tomato by Pseudomonas aeruginosa 7NSK2. Appl. Environ. Microbiol. 1996, 62, 865–871.
- Gaille, C.; Kast, P.; Haas, D. Salicylate biosynthesis in Pseudomonas aeruginosa: Purification and characterization of PchB, a novel bifunctional enzyme displaying isochorismate pyruvate-lyase and chorismate mutase activities. J. Biol. Chem. 2002, 277, 21768–21775.
- Meneely, K.M.; Luo, Q.; Riley, A.P.; Taylor, B.; Roy, A.; Stein, R.L.; Prisinzano, T.E.; Lamb, A.L. Expanding the results of a high throughput screen against an isochorismate-pyruvate lyase to enzymes of a similar scaffold or mechanism. Bioorg. Med. Chem. 2014, 22, 5961–5969.
- Chen, C.; Bélanger, R.R.; Benhamou, N.; Paulitz, T.C. Role of salicylic acid in systemic resistance induced by Pseudomonas spp. against Pythium aphanidermatum in cucumber roots. Eur. J. Plant Pathol. 1999, 105, 477–486.
- Saikia, R.; Singh, T.; Kumar, R.; Srivastava, J.; Srivastava, A.K.; Singh, K.; Arora, D.K. Role of salicylic acid in systemic resistance induced by Pseudomonas fluorescens against Fusarium oxysporum f. sp. ciceri in chickpea. Microbiol. Res. 2003, 158, 203–213.
- Saikia, R.; Srivastava, A.K.; Singh, K.; Arora, D.K.; Lee, M.-W. Effect of iron availability on induction of systemic resistance to fusarium wilt of chickpea by Pseudomonas spp. Mycobiology 2005, 33, 35–40.
- Saikia, R.; Kumar, R.; Arora, D.K.; Gogoi, D.K.; Azad, P. Pseudomonas aeruginosa inducing rice resistance against Rhizoctonia solani: Production of salicylic acid and peroxidases. Folia Microbiol. 2006, 51, 375–380.
- Sokol, P.A.; Lewis, C.J.; Dennis, J.J. Isolation of a novel siderophore from Pseudomonas cepacia. J. Med. Microbiol. 1992, 36, 184–189.
- Islam, M.; Ali, M.; Choi, S.-J.; Park, Y.-I.; Baek, K.-H. Salicylic acid-producing endophytic bacteria increase nicotine accumulation and resistance against wildfire disease in tobacco plants. Microorganisms 2020, 8, 31.
- Hertlein, G.; Müller, S.; Garcia-Gonzalez, E.; Poppinga, L.; Süssmuth, R.D.; Genersch, E. Production of the catechol type siderophore bacillibactin by the honey bee pathogen Paenibacillus larvae. PLoS ONE 2014, 9, e108272.
- Saxena, B.; Modi, M.; Modi, V. V Isolation and characterization of siderophores from Azospirillum lipoferum D-2. Microbiology 1986, 132, 2219–2224.
- Lee, J.Y.; Passalacqua, K.D.; Hanna, P.C.; Sherman, D.H. Regulation of petrobactin and bacillibactin biosynthesis in Bacillus anthracis under iron and oxygen variation. PLoS ONE 2011, 6, e20777.
- Barbeau, K.; Zhang, G.; Live, D.H.; Butler, A. Petrobactin, a photoreactive siderophore produced by the oil-degrading marine bacterium Marinobacter hydrocarbonoclasticus. J. Am. Chem. Soc. 2002, 124, 378–379.
- Forchetti, G.; Masciarelli, O.; Izaguirre, M.J.; Alemano, S.; Alvarez, D.; Abdala, G. Endophytic bacteria improve seedling growth of sunflower under water stress, produce salicylic acid, and inhibit growth of pathogenic fungi. Curr. Microbiol. 2010, 61, 485–493.
- Kesaulya, H.; Hasinu, J.; Tuhumury, G. Potential of Bacillus spp. produces siderophores insuppressing thewilt disease of banana plants. IOP Conf. Ser. Earth Environ. Sci. 2018, 102, 12016.
- Nandhini, S.; Sendhilvel, V.; Babu, S. Endophytic bacteria from tomato and their efficacy against Fusarium oxysporum f.sp. lycopersici, the wilt pathogen. J. Biopestic. 2012, 5, 178–185.
- Ciche, T.A.; Blackburn, M.; Carney, J.R.; Ensign, J.C. Photobactin: A catechol siderophore produced by Photorhabdus luminescens, an entomopathogen mutually associated with Heterorhabditis bacteriophora NC1 nematodes. Appl. Environ. Microbiol. 2003, 69, 4706–4713.
- Xie, F.; Dai, S.; Shen, J.; Ren, B.; Huang, P.; Wang, Q.; Liu, X.; Zhang, B.; Dai, H.; Zhang, L. A new salicylate synthase AmS is identified for siderophores biosynthesis in Amycolatopsis methanolica 239T. Appl. Microbiol. Biotechnol. 2015, 99, 5895–5905.
- Müller, S.I.; Valdebenito, M.; Hantke, K. Salmochelin, the long-overlooked catecholate siderophore of Salmonella. BioMetals 2009, 22, 691–695.
- Crouch, M.-L.V.; Castor, M.; Karlinsey, J.E.; Kalhorn, T.; Fang, F.C. Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2008, 67, 971–983.
- Hantke, K.; Nicholson, G.; Rabsch, W.; Winkelmann, G. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. USA 2003, 100, 3677–3682.
- Griffiths, G.L.; Sigel, S.P.; Payne, S.M.; Neilands, J.B. Vibriobactin, a siderophore from Vibrio cholerae. J. Biol. Chem. 1984, 259, 383–385.
- Thode, S.K.; Rojek, E.; Kozlowski, M.; Ahmad, R.; Haugen, P. Distribution of siderophore gene systems on a Vibrionaceae phylogeny: Database searches, phylogenetic analyses and evolutionary perspectives. PLoS ONE 2018, 13, e0191860.
- Simpson, L.M.; Oliver, J.D. Siderophore production by Vibrio vulnificus. Infect. Immun. 1983, 41, 644–649.
- Okujo, N.; Saito, M.; Yamamoto, S.; Yoshida, T.; Miyoshi, S.; Shinoda, S. Structure of vulnibactin, a new polyamine-containing siderophore from Vibrio vulnificus. Biometals 1994, 7, 109–116.
- Tan, W.; Verma, V.; Jeong, K.; Kim, S.Y.; Jung, C.-H.; Lee, S.E.; Rhee, J.H. Molecular characterization of vulnibactin biosynthesis in Vibrio vulnificus indicates the existence of an alternative siderophore. Front. Microbiol. 2014, 5, 1.
- Pfeifer, B.A.; Wang, C.C.C.; Walsh, C.T.; Khosla, C. Biosynthesis of yersiniabactin, a complex polyketide-nonribosomal peptide, using Escherichia coli as a heterologous host. Appl. Environ. Microbiol. 2003, 69, 6698–6702.
- Kerbarh, O.; Ciulli, A.; Howard, N.I.; Abell, C. Salicylate biosynthesis: Overexpression, purification, and characterization of Irp9, a bifunctional salicylate synthase from Yersinia enterocolitica. J. Bacteriol. 2005, 187, 5061–5066.
- Perry, R.D.; Fetherston, J.D. Yersiniabactin iron uptake: Mechanisms and role in Yersinia pestis pathogenesis. Microbes Infect. 2011, 13, 808–817.
- Paauw, A.; Leverstein-van Hall, M.A.; van Kessel, K.P.M.; Verhoef, J.; Fluit, A.C. Yersiniabactin reduces the respiratory oxidative stress response of innate immune cells. PLoS ONE 2009, 4, e8240.
- Quadri, L.E.N.; Sello, J.; Keating, T.A.; Weinreb, P.H.; Walsh, C.T. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 1998, 5, 631–645.
- De Voss, J.J.; Rutter, K.; Schroeder, B.G.; Su, H.; Zhu, Y.; Barry, C.E., 3rd. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. USA 2000, 97, 1252–1257.
- Harrison, A.J.; Yu, M.; Gårdenborg, T.; Middleditch, M.; Ramsay, R.J.; Baker, E.N.; Lott, J.S. The structure of MbtI from Mycobacterium tuberculosis, the first enzyme in the biosynthesis of the siderophore mycobactin, reveals it to be a salicylate synthase. J. Bacteriol. 2006, 188, 6081–6091.
- Sritharan, M. Iron homeostasis in Mycobacterium tuberculosis: Mechanistic insights into siderophore-mediated Iron uptake. J. Bacteriol. 2016, 198, 2399–2409.
- Patel, K.; Butala, S.; Khan, T.; Suvarna, V.; Sherje, A.; Dravyakar, B. Mycobacterial siderophore: A review on chemistry and biology of siderophore and its potential as a target for tuberculosis. Eur. J. Med. Chem. 2018, 157, 783–790.
- Shyam, M.; Shilkar, D.; Verma, H.; Dev, A.; Sinha, B.N.; Brucoli, F.; Bhakta, S.; Jayaprakash, V. The Mycobactin biosynthesis pathway: A prospective therapeutic target in the battle against tuberculosis. J. Med. Chem. 2020, 64, 71–100.
- Ho, Y.-N.; Lee, H.-J.; Hsieh, C.-T.; Peng, C.-C.; Yang, Y.-L. Chemistry and biology of salicyl-capped siderophores. In Studies in Natural Products Chemistry; Atta-ur-Rahman, B.T.-S., Ed.; Elsevier: Amsterdam, The Netherlands, 2018; Volume 59, pp. 431–490. ISBN 1572-5995.
- Masschelein, J.; Mattheus, W.; Gao, L.-J.; Moons, P.; Van Houdt, R.; Uytterhoeven, B.; Lamberigts, C.; Lescrinier, E.; Rozenski, J.; Herdewijn, P.; et al. A PKS/NRPS/FAS hybrid gene cluster from Serratia plymuthica RVH1 encoding the biosynthesis of three broad spectrum, Zeamine-related antibiotics. PLoS ONE 2013, 8, e54143.
- Süssmuth, R.D.; Mainz, A. Nonribosomal peptide synthesis—Principles and prospects. Angew. Chem. Int. Ed. 2017, 56, 3770–3821.
- Wang, H.; Fewer, D.P.; Holm, L.; Rouhiainen, L.; Sivonen, K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc. Natl. Acad. Sci. USA 2014, 111, 9259–9264.
- Jaremko, M.J.; Davis, T.D.; Corpuz, J.C.; Burkart, M.D. Type II non-ribosomal peptide synthetase proteins: Structure, mechanism, and protein–protein interactions. Nat. Prod. Rep. 2020, 37, 355–379.
- Bloudoff, K.; Schmeing, T.M. Structural and functional aspects of the nonribosomal peptide synthetase condensation domain superfamily: Discovery, dissection and diversity. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 1587–1604.
- Sayari, M.; van der Nest, M.A.; Steenkamp, E.T.; Soal, N.C.; Wilken, P.M.; Wingfield, B.D. Distribution and evolution of nonribosomal peptide synthetase gene clusters in the Ceratocystidaceae. Genes 2019, 10, 328.
- Walsh, C.T.; Chen, H.; Keating, T.A.; Hubbard, B.K.; Losey, H.C.; Luo, L.; Marshall, C.G.; Miller, D.A.; Patel, H.M. Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Curr. Opin. Chem. Biol. 2001, 5, 525–534.
- White-Stevens, R.H.; Kamin, H. Studies of a flavoprotein, salicylate hydroxylase: I. Preparation, properties, and the uncoupling of oxygen reduction from hydroxylation. J. Biol. Chem. 1972, 247, 2358–2370.
- Costa, D.M.A.; Gómez, S.V.; de Araújo, S.S.; Pereira, M.S.; Alves, R.B.; Favaro, D.C.; Hengge, A.C.; Nagem, R.A.P.; Brandão, T.A.S. Catalytic mechanism for the conversion of salicylate into catechol by the flavin-dependent monooxygenase salicylate hydroxylase. Int. J. Biol. Macromol. 2019, 129, 588–600.
- You, I.S.; Ghosal, D.; Gunsalus, I.C. Nucleotide sequence analysis of the Pseudomonas putida PpG7 salicylate hydroxylase gene (nahG) and its 3′-flanking region. Biochemistry 1991, 30, 1635–1641.
- Bosch, R.; Moore, E.R.B.; García-Valdés, E.; Pieper, D.H. NahW, a novel, inducible salicylate hydroxylase involved in mineralization of naphthalene by Pseudomonas stutzeri AN10. J. Bacteriol. 1999, 181, 2315–2322.
- Pozdnyakova-Filatova, I.; Petrikov, K.; Vetrova, A.; Frolova, A.; Streletskii, R.; Zakharova, M. The naphthalene catabolic genes of Pseudomonas putida BS3701: Additional regulatory control. Front. Microbiol. 2020, 11, 1217.
- Maspoli, A.; Wenner, N.; Mislin, G.L.A.; Reimmann, C. Functional analysis of pyochelin-/enantiopyochelin-related genes from a pathogenicity island of Pseudomonas aeruginosa strain PA14. BioMetals 2014, 27, 559–573.
- Ronnebaum, T.A.; Lamb, A.L. Nonribosomal peptides for iron acquisition: Pyochelin biosynthesis as a case study. Curr. Opin. Struct. Biol. 2018, 53, 1–11.
- Schalk, I.J.; Rigouin, C.; Godet, J. An overview of siderophore biosynthesis among fluorescent Pseudomonads and new insights into their complex cellular organization. Environ. Microbiol. 2020, 22, 1447–1466.
- Wyckoff, E.E.; Smith, S.L.; Payne, S.M. VibD and VibH are required for late steps in vibriobactin biosynthesis in Vibrio cholerae. J. Bacteriol. 2001, 183, 1830–1834.
- Lawlor, M.S.; O’connor, C.; Miller, V.L. Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect. Immun. 2007, 75, 1463–1472.
- Manos-Turvey, A.; Bulloch, E.M.M.; Rutledge, P.J.; Baker, E.N.; Lott, J.S.; Payne, R.J. Inhibition studies of Mycobacterium tuberculosis salicylate synthase (MbtI). ChemMedChem 2010, 5, 1067–1079.
- Miethke, M.; Marahiel, M.A. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451.
- Hider, R.C.; Kong, X. Chemistry and biology of siderophores. Nat. Prod. Rep. 2010, 27, 637–657.
- Golonka, R.; San Yeoh, B.; Vijay-Kumar, M. The iron tug-of-war between bacterial siderophores and innate immunity. J. Innate Immun. 2019, 11, 249–262.
- Loba, V.C.; Pollmann, S. Highly sensitive salicylic acid quantification in milligram amounts of plant tissue. In Plant Hormones: Methods and Protocols; Kleine-Vehn, J., Sauer, M., Eds.; Springer: New York, NY, USA, 2017; pp. 221–229. ISBN 978-1-4939-6469-7.
- Schmelz, E.A.; Engelberth, J.; Alborn, H.T.; O’ Donnell, P.; Sammons, M.; Toshima, H.; Tumlinson, J.H. Simultaneous analysis of phytohormones, phytotoxins, and volatile organic compounds in plants. Proc. Natl. Acad. Sci. USA 2003, 100, 10552–10557.
- Huang, Z.; Wang, Z.; Shi, B.; Wei, D.; Chen, J.; Wang, S.; Gao, B. Simultaneous determination of salicylic acid, jasmonic acid, methyl salicylate, and methyl jasmonate from Ulmus pumila Leaves by GC-MS. Int. J. Anal. Chem. 2015, 2015, 698630.
- Allasia, V.; Ponchet, M.; Quentin, M.; Favery, B.; Keller, H. Quantification of salicylic acid (SA) and SA-glucosides in Arabidopsis thaliana. Bio-Protocol 2018, 8, e2844.
- Louden, B.C.; Haarmann, D.; Lynne, A.M. Use of blue agar CAS assay for siderophore detection. J. Microbiol. Biol. Educ. 2011, 12, 51–53.
- Alexander, D.B.; Zuberer, D.A. Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 1991, 12, 39–45.
- Arnow, L.E. Colorimetric determination of the components of 3, 4-dihydroxyphenylalanine-tyrosine mixtures. J. Biol. Chem. 1937, 118, 531–537.




