1. SA
Phenolic compounds contain an aromatic benzene ring with one or more hydroxyl groups produced as secondary metabolites in nature, primarily in plants and some microorganisms [
1,
2]. They were presumed to be the byproducts of metabolic pathways, and dispensable for important processes common to all organisms [
3]. While plants and a few microorganisms (especially bacteria and fungi) produce phenolics, there are variations between and within species [
4]. In plants, phenolics play crucial roles in the regulation of different biochemical and physiological processes [
5]. One such important phenolic compound is 2-hydroxy benzoic acid, called salicylic acid (SA) [
6,
7]. There is a wide variation in the basal levels of SA, with up to 100-fold differences among plants of the same family, and significant differences between species [
8,
9].
Although SA or its related metabolites have long been used as a pain reliever, the active extract of the inner bark of the
Salix alba was isolated and named ‘salicin’ by a German chemist, Johann Buchner, in 1828 [
10]. However, the first report of SA signaling in plants was published in 1987, when a mass spectroscopic analysis of the male flowers of calla lily indicated their role in heat production [
11]. Subsequent studies revealed that the reproductive organ of gymnosperms and angiosperms displays a thermogenesis phenomenon due to SA signaling [
8,
12]. A few years after discovering its role in thermogenesis, SA has emerged as a signaling molecule during pathogen infection [
13]. Exogenous SA application into tobacco plant leaves was shown to induce pathogenesis-related proteins and improve resistance to tobacco mosaic virus (TMV) infections [
14,
15]. Since then, many research groups have demonstrated that the increased levels of SA are associated with the induction of defense genes and systemic acquired resistance in the plant species [
16,
17]. Afterwards, many studies have established that SA is a key signal molecule in regulating the activation of local and systemic defense responses against infections by pathogens [
12,
16,
18]. With SA being accepted as the ‘sixth’ principal plant hormone in the late 1990s [
19], several scientific groups started working on its different physiological roles, barring thermogenesis and plant immunity [
16,
20,
21,
22,
23]. In a short period of time, SA has become an essential signaling molecule in plants and plays a regulatory role in abiotic stresses, like heat stress and drought, and biotic stresses, such as the systemic acquired resistance mediated defense response against pathogen infection [
24,
25,
26,
27,
28,
29]. In addition, SA’s function also influences plant growth and development by regulating various processes, such as photosynthesis, respiration, vegetative growth, seed germination, flowering, senescence, etc. [
30,
31,
32,
33]. Furthermore, acetylsalicylic acid (under the popular trade name, Aspirin) has been an important agent for treating various medical conditions [
34]. It has been widely used to manage fever and pain, as well as for the management of cardiovascular diseases [
34,
35,
36] and dermatological conditions such as acne, blisters and pruritus [
37].
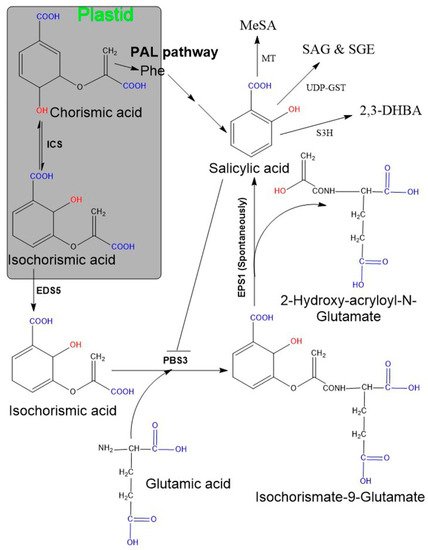
Chorismate (the end-product of the shikimate pathway) is employed as a primary source for the SA biosynthetic pathway. The shikimate pathway starts with erythrose-4-phosphate and phosphoenolpyruvate, and a series of condensation/redox reactions occur, resulting in the formation of chorismate [
38,
39]. This exclusively conserved pathway is found in bacteria, plants, and fungi, but not in animals [
40]. Chorismate is a central metabolic route for the biosynthesis of aromatic amino acids (L-tryptophan, L-phenylalanine, and L-tyrosine) and various aromatic secondary metabolites (such as alkaloids, flavonoids, lignins, aromatic antibiotics, and SA) [
41,
42]. Thus, chorismate acts as a common connecting precursor of primary and secondary metabolism [
43,
44]. The first step in the SA biosynthesis is to convert chorismate to isochorismate by using isochorismate synthase (ICS), or its homologous enzyme, common to both bacteria and plants [
20,
45]. Seven chorismate-utilizing enzymes exist. Of these, five (ICS, salicylate synthase (SAS), anthranilate synthase, aminodeoxyisochorismate synthase, aminodeoxychorismate synthase) are structural homologues and are collectively known as menaquinone, siderophore, and tryptophan (MST) enzymes [
46]. The others, two chorismate-utilizing enzymes (chorismate mutase and chorismate lyase), belong to the non-MST family and are responsible for biosynthesizing phenylalanine, tyrosine amino acid, and ubiquinone. Among the above-mentioned seven enzymes, five are present in both plants and bacteria, while the other two (SAS and chorismate lyase) are exclusively reported in bacteria.
The biosynthesis of SA and its functions in plants have now been completely understood through two breakthrough studies [
47,
48]. However, knowledge on microbial SA biosynthesis is still scarce, and their functions are yet to be elucidated. In plants, SA functions as a hormone, regulating several physiological processes, such as biotic and abiotic stress responses, seed germination, and flowering [
6,
24,
25]. Meanwhile, in bacteria, SA is mainly associated with salicyl-derived siderophores [
49,
50], and some of its derivative compounds also act as antibacterial agents, e.g., promysalin [
51]. Salicyl-derived siderophores are produced by few plant-growth-promoting rhizobacteria and pathogenic bacteria [
52,
53]. Considering the practical importance of SA, the objective of this study is to investigate the occurrence of bacterial salicylate, its biosynthetic pathways, and compare with plant SA biosynthesis.
3. SA-Derived (Catecholate) Siderophores in Bacteria
Siderophores are small Fe
3+-chelating secondary metabolites secreted by bacteria and fungi under low-iron conditions [
68]. In addition, some plants also produce siderophores for iron acquisition [
69]. Generally, siderophores are produced intracellularly and secreted outside the cell as iron-free (deferri) compounds. After scavenging iron (Fe
3+ form), the iron-siderophore complex is transported into the cell by the transport system. Based on their structural features and iron-chelating functional groups, siderophores have been classified into three main classes: catecholate (also termed as pheno-catecholate or salicyl-derived siderophore), hydroxamate, and carboxylate [
70]. In addition, there are mixed-types (containing more than one of the above-named moieties). In the aforementioned three major classes of siderophores, catecholate siderophores are exclusively produced by bacteria. Usually, bacterial SA is assimilated into the salicyl-derived (termed as salicylates) siderophore backbone. A few mycorrhizal fungi (
Ustilago maydis and
Phialocephala fortinii) have also been reported to produce hydroxamate siderophore [
71]. Further, the
Pseudomonas spp. have been documented as the main synthesizer for the catecholate siderophore. More than 100 salicylate siderophores have been reported, and a few are listed in . Based on their major structural moieties, the catecholate siderophores can be classified into three classes, oxazoline/oxazole, thiazoline/thiazole, and serine-backbone groups. All catecholate siderophores use SA or its hydroxylated derivate, 2,3-dihydroxybenzoic acid (2,3-DHBA), as the common precursor, and its biosynthesis has been well established to involve enzymatic transformations starting from chorismate [
72]. Furthermore, several bacteria are capable of producing more than one type of siderophore, such as mycobacterium (produces both types, catecholate and carboxylate).
Table 1. Salicylic acid-producing bacteria and their salicylate-derived siderophores.
| Bacteria Species |
Salicylate-Siderophore |
Bacterial Source |
NRPS Biosynthetic Genes |
References |
| Anabaena cylindrical # |
Anachelin |
Pond |
ICS-IPL |
[73,74] |
| Acinetobacter baumannii |
Acinetobactin * |
Human |
ICS-IPL |
[75,76] |
| Pseudomonas fluorescens CHA0 |
Pyochelin |
Tobacco rhizosphere |
ICS-IPL |
[77,78,79] |
| P. fluorescens WCS374 |
Pyochelin |
Potato rhizosphere |
ICS-IPL |
[80,81] |
| P. fluorescens WCS417 |
Pyochelin |
Wheat rhizosphere |
ICS-IPL |
[80,82,83] |
| Serratia marcescens |
Pyochelin |
Cucumber/Tobacco rhizosphere |
ICS-IPL |
[84,85] |
| P. aeruginosa 7NSK2 |
Pyochelin |
Barley roots |
ICS-IPL |
[78,86,87,88] |
| P. aureofaciens 63-28 |
Pyochelin |
Cucumber roots |
ICS-IPL |
[89] |
| P. corrugata 13 |
Pyochelin |
Cucumber roots |
ICS-IPL |
[89] |
| P. fluorescens Pf4–92 |
Pseudobactin * |
Chickpea rhizosphere |
ICS-IPL |
[90,91] |
| P. fluorescens PICF3 |
Pyochelin |
Olive root |
ICS-IPL |
[81] |
| P. aeruginosa RsG18 and P. aeruginosa RsG27 |
Pseudobactin * |
Rhizosphere soil |
ICS-IPL |
[92] |
| P. cepacia |
Azurechelin * |
Human |
ICS-IPL |
[93] |
| P.tremae |
N/A |
Leaves of Salix babylonica |
N/A |
[94] |
| Paenibacillus larvae |
Bacillibactin |
Larvae of honeybees |
ICS-IPL |
[95] |
| Azospirillum iipoferurn D-2 |
N/A |
Digireria roots |
ICS-IPL |
[96] |
| Bacillus anthracis |
Petrobactin |
Sunflower soil |
ICS-IPL |
[97] |
| Marinobacter hydrocarbonoclasticus |
Petrobactin |
N/A |
ICS-IPL |
[98] |
| B. pumilus SF3 |
Bacillibactin |
Sunflower plant |
ICS-IPL |
[95,99,100] |
| B. subtilis |
Bacillibactin |
Banana plant |
|
[100] |
| Citrobacter |
Enterobactin |
Tomato roots |
ICS-IPL |
[101] |
| Klebsiella pneumoniae |
Enterobactin |
Tomato leaves |
ICS-IPL |
[101] |
| Photorhabdus luminescens |
Photobactin |
Nematode(Heterorhabditis bacteriophora) |
SAS |
[102] |
| Amycolatopsis methanolica 239T |
Amychelin |
Soil |
SAS |
[103] |
| Salmonella enterica serotype Typhimurium |
Salmochelin |
Human |
SAS |
[104,105,106] |
| Vibrio cholerae |
Vibriobactin |
Human |
SAS |
[107,108] |
| V. vulnificus |
Vulnibactin |
Human |
SAS |
[108,109,110,111] |
| Yersinia enterocolitica |
Yersiniabactin * |
Human |
SAS |
[112,113,114] |
| Y. pestis |
Yersiniabactin * |
Human |
SAS |
[115] |
| Mycobacterium tuberculosis |
Mycobactin |
Human |
SAS |
[116,117,118,119,120,121] |
Siderophore biosynthesis occurs in two ways: the nonribosomal peptide synthetase and polyketide synthases (NRPS/PKSs) pathway, and the NRPS-independent siderophore (NIS) synthetase pathway. Both NRPS/PKSs and NIS biosynthetic enzymes are encoded as BGCs on the microbial genome. Further, each gene is encoded by a specific module and represents a specific enzyme. Bacteria synthesize salicyl-derived siderophores by two methods: either NRPS biosynthetic gene clusters (NRPS BGC) or NRPS/PKS hybrid biosynthetic gene clusters (NRPS/PKS BGC). For example, pyochelin and bacillibactin biosynthesis involves NRPS, while yersiniabactin and mycobactin involve both NRPS/PKS [
122]. PKSs are commonly found in fungi, but they are also contained in a few bacteria. Additionally, some bacteria employ other gene clusters along with NRPS/PKS biosynthetic gene clusters [
123].
NRPS catalyzes the synthesis of highly diverse natural microbial products, such as antibiotics, toxins, and siderophores [
124]. They are found in all three domains of life (bacteria, archaea, and eukarya) and can synthesize a peptide from a variety of standard, non-proteinogenic amino acids, as well as carboxylic and hydroxy acids. Typically, NRPSs are multimodular enzymes consisting of repeated modules (type I NRPS), but nonmodular enzymes (type II NRPS) are also reported [
125,
126]. In multimodular NRPS, each module contains three domains: adenylation domain, thiolation domain (also termed as peptidyl carrier protein), and condensation domain. Condensation domains of NRPS (which catalyze the amide bond formation) are functionally the most important and suitable target for natural product genomic analysis [
125,
127]. Nonmodular NRPSs are linear, comprising specialized tailoring enzymes for more diversification. Nonmodular NRPSs commonly combine their substrate with other pathways to generate a final product [
126]. All NRPS biosynthetic genes are organized as operons in BGCs, and their regulation takes place at a transcriptional or post-translational level [
128,
129]. Additionally, salicylate-derived siderophores comprising BGC show significant similarity among the same bacterial genus. For instance, the salicylate-coding gene cluster of
P. fluorescens was compared with the other three
Pseudomonas strains and showed a 99% (having 100% query coverage) identity in conserved biosynthetic gene sequences [
56].
Biosynthetic genes are encoded by a specific module of the NRPSs gene cluster. Two SA biosynthetic routes have been illustrated in bacteria, and the SA is finally incorporated into salicyl-derived siderophores or other metabolites (). First, most SA-producing bacterial spp. (e.g.,
Pseudomonas and
Bacillus spp.) operate an ICS-like biosynthetic pathway. Here, the first step is converting chorismate to its isomer, isochorismate, by the ICS enzyme, while the next step requires another enzyme, isochorismate pyruvate lyase (IPL). IPL converts isochorismate to SA and pyruvate, and is considered a crucial enzyme for bacterial salicylate biosynthesis [
45]. So far, no orthologous gene corresponding to bacterial IPL has been identified in plant genomes [
7]. Second, some pathogenic bacterial spp. (
Yersinia,
Vibrio,
Salmonella, and
Mycobacterium) possess a single bifunctional enzyme called salicylate synthase (SAS) that can directly convert chorismate to SA through an isochorismate intermediate. All bacterial biosynthetic genes,
ICS,
IPL, and
SAS are encoded by a specific module of an NRPS BGC. Further, both ICS and SAS enzymes evolutionally originate from a common ancestor and are highly similar to each other [
46,
113]. The type I NRPS and the type II NRPS siderophores are represented by the vibriobactin siderophore (in
Vibrio) and mycobactin (in
Mycobacterium), respectively.
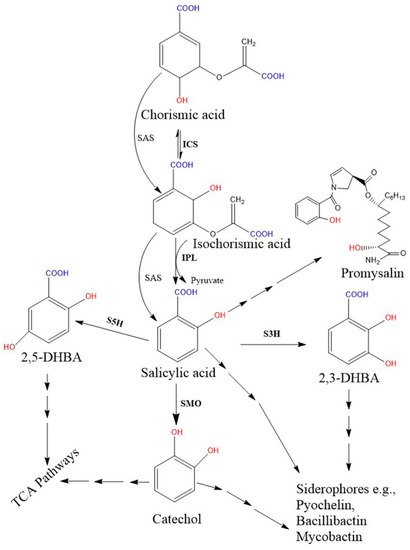
Figure 2. Proposed bacterial pathway for the biosynthesis of SA and their derivative catecholate siderophore. The catecholate siderophore group is only found in bacteria, and their core structures are synthesized from the SA-BGC system. In the
Pseudomonas spp., SA is synthesized from chorismate via a two-step enzymatic process (
pchA; ICS and
pchB; IPL), while the single bifunctional enzyme SAS encoded by
MbtI (in Mycobacterium) and
Irp9 (in Yersinia) converts chorismate to SA. Further, SMO (also called S1H) converts SA to 1,2-dihydroxybenzene (catechol). Additionally, hydroxylation of SA results in 2,3-DHBA and 2,5 DHBA. Here, salicylate/2, 3-DHBA/catechol can be converted to salicyl-derived siderophores with the help of more than one enzyme. Moreover, catechol and gentisate (2, 5-DHBA) are also metabolized in the TCA pathway. Promysalin, an antibiotic synthesized from SA is also reported in a few
Pseudomonas spp. [
51]. Solid arrows denote single enzymatic steps, while several arrows denote multiple sequential enzymatic steps. All the demonstrated chemical structures are drawn using the ChemDraw 13.0 software (PerkinElmer, Waltham, MA, USA). Abbreviations: ICS, isochorismate synthase; IPL, isochorismate pyruvate lyase; SAS, salicylate synthase; SMO, salicylate1-monooxygenase; S1H, salicylate 1-hydroxylase; S3H, SA 3-hydroxylase; S5H, salicylate 5-hydroxylase; DHBA, dihydroxybenzoic acid; TCA, tricarboxylic acid.
Salicylate monooxygenase (also known as salicylate 1-hydroxylase) converts salicylate into catechol (1, 2-dihydroxybenzene). This enzyme is also important for salicyl-derived siderophore synthesis and is encoded by a specific module of the NRPS biosynthetic gene clusters [
129,
130,
131]. These enzymes in the
Pseudomonas species are encoded from the genes of
NahG in
P. fluorescens 142 NF and
P. putida [
131,
132],
NahW in
P. stutzeri AN10 [
133], and
NahU in
P. putida BS3701 [
134].
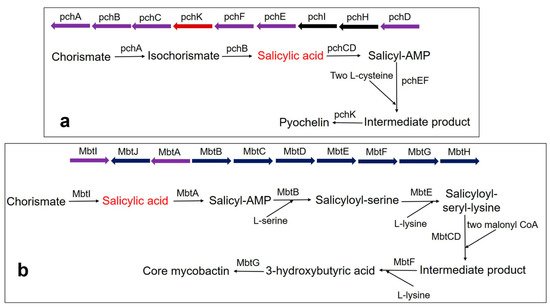
Pyochelin siderophore is made from one molecule of SA and two molecules of cysteine by a thiotemplate mechanism. In
Pseudomonas aeruginosa, the biosynthesis of pyochelin takes place by the BGC comprising pchDHIEFKCBA operon [
135,
136,
137]. Two enzymes, pchA (ICS) and pchB (IPL), start the synthesis. Chorismate is converted to isochorismate by the pchA and subsequently, the isochorismate is converted to salicylate by pchB. Subsequently, salicylate is activated by pchD (salicyl-adenylating enzyme). In the meantime, enzyme pchC (encoded for type II thioesterase) removes wrongly charged molecules. Afterward, two tailoring enzymes (pchE and pchF) add L-cysteine residues and perform cyclization and epimerization, resulting in the formation of an intermediate product. Finally, the action of pchK (saccharopine reductase) results in pyochelin synthesis (a).
Figure 3. Biosynthesis pathway of catecholate siderophore in bacteria (a) Pyochelin biosynthesis in P. aeruginosa using BGC comprising the pchDHIEFKCBA operon. The biosynthesis of pyochelin is initiated from chorismate by two salicylic acid synthesizing enzymes pchA (ICS) and pchB (IPL), encoded by NRPS BGC. Other gene encoding enzymes are pchC (type II thioesterase); pchD (salicyl-adenylating enzyme); pchE and pchF (tailoring enzyme that adds L-cysteine and performs cyclization and epimerization); pchK (saccharopine reductase); pchH and pchI (transporter enzyme). (b) Core mycobactin scaffold biosynthesis in M. tuberculosis by using BGC comprising the MbtA-J operon. Biosynthesis is carried out by a cluster consisting of 10 essential gene encodings for the enzyme (MbtA-MbtJ). Conversion of chorismate to salicylate requires the main enzyme, MbtI. MbtI (salicylate synthase); MbtA (salicyl-adenylating enzyme); MbtB (enzyme for acyl carrier protein); MbtB and MbtE (enzyme for the addition of serine and lysine residues, respectively); MbtC & MbtD (enzyme for the addition of malonyl CoA residues); MbtF (enzyme for the addition of terminal lysine); MbtG (enzyme for N6-hydroxylation of L-lys); MbtJ and MbtH (enzyme encoding unknown functions).
Mycobactin biosynthesis is performed by an NRPS/PKS hybrid system in
M. tuberculosis. The biosynthetic genes are encoded by two BGCs (Mbt-1 and Mbt-2) and include 14 gene encoding enzymes [
121]. The Mbt-1 cluster (
mbtA-mbtJ) consists of 10 essential gene encodings for biosynthetic enzymes (MbtA-MbtJ) involved in core mycobactin scaffold formation (b), while Mbt-2 includes four genes encoding for acyltransferase. Mbt-1 consists of salicylate synthase encoded as
Mbt I, which initiates the synthesis of mycobactin followed by the activation of salicylic acid by the adenylating enzyme, MbtA. Similarly,
V. cholerae produces the catechol siderophore, vibriobactin, using vibABCDEFH BGC operon. Here, the BGC operon contains SAS encoded by
VibH [
108,
138].
In several bacteria, siderophores can act as virulence factors to combat the host immune system [
139,
140]. Many catecholate-type siderophores are also found in pathogenic bacteria, such as mycobactin (
M. tuberculosis), vibriobactin (
Vibrio cholera), salmochelin (
Salmonella enterica), petrobactin (
Bacillus anthracis), and Yersiniabactin (
Yersinia enterocolitica), and are responsible for their pathogenicity ( [
53,
141]. Among all the categories of siderophores, the catecholate-type has stronger iron affinities than transferrin and lactoferrin [
142]. These features help it to gain a high degree of pathogenicity in catecholate-producing pathogenic bacteria. Pathogens sequester iron from the host protein using siderophores. To counter this, lipocalin-2 protein is released by the host immune system, which sequesters the catecholate-type siderophores and thus impedes bacterial growth [
53,
143]. Furthermore, few bacteria, such as
B. anthracis and
S. enterica, secrete lipocalin-2 resistant stealth siderophores and evade the host immune system. Hence, siderophores and their biosynthetic enzymes (ICS/IPL/SAS) could act as a suitable target for drugs and could be helpful for medicine development in the future.