
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Emma Wilkinson | + 1031 word(s) | 1031 | 2021-04-25 06:07:52 | | | |
| 2 | Catherine Yang | Meta information modification | 1031 | 2021-05-14 02:56:49 | | |
Video Upload Options
RNA modifications are diverse post-transcriptional modifications that regulate RNA metabolism and gene expression. RNA modifications, and the writers, erasers, and readers that catalyze these modifications, serve as important signaling machineries in cellular stress responses and disease pathogenesis.
1. Introduction
RNA modifications are covalent chemical modifications of RNA molecules. To date, over 100 chemical modifications of RNA species have been identified [1][2]. The regulation and function of RNA modifications have recently emerged as pivotal mechanisms that regulate a wide range of biological and pathological processes, giving rise to the field known as epitranscriptomics.
RNA modifications are regulated through the coordination of ‘writers’, ‘erasers’ and ‘readers’, which deposit, remove, and recognize RNA modifications, respectively (Figure 1A). These enzymes represent key elements in patterning the epitranscriptomic landscape.
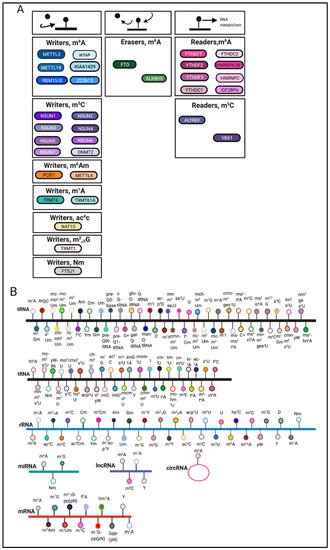
RNA modifications can occur on various RNA species including mRNA, tRNA, rRNA and other non-coding RNAs [9] (Figure 1B). Among these RNA species, transfer RNAs (tRNAs) contain the most RNA modifications [10]. One of the most abundant modifications on tRNA and rRNA is 5-methylcytosine (m5C) [11]. In comparison, mRNA modifications were more difficult to identify and characterize due to their low-abundance. The advent of sophisticated sequencing technologies and methods has generated a renewed interest in mRNA modifications.
2. RNA Modifications in Diseases
2.1. Cancer
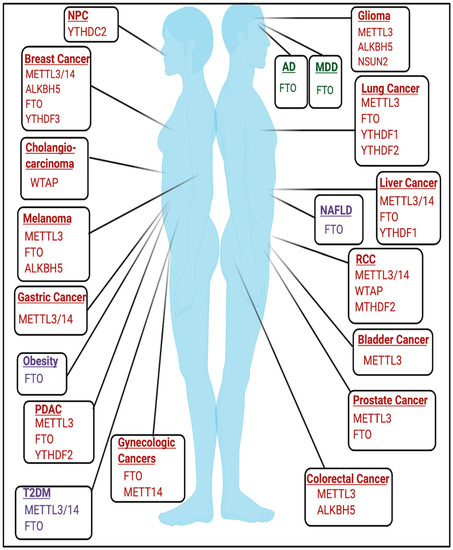
2.2. Developmental and Neurologic Disorders
The role of RNA modifications in the context of developmental and neurological disorders remains an active area of study. m6A has been previously found to play important roles in embryonic development and neurobiological functions [25][26]. The roles of m6A and other RNA modifications in mediating neurologic function are further discussed elsewhere [25][27][28][29][30].
The necessity of m6A in development is emphasized by early embryonic lethality in mettl3 KO mice [31]. Conditional mettl3 knockout in murine brains also resulted in severe developmental defects within the cerebrum and cortex and induced apoptosis in cerebella granule cells (CGCs) [32]. FTO may also be important in mediating development as expression of catalytically inactive mutant FTO(R316Q) resulted in severe growth defects [33]. Mutations in tRNA methyltransferases have been implicated in developmental disorders and are detailed in [34]. NSUN2 mutations have been linked to microcephaly, intellectual disability, and Dubowitz Syndrome, which is characterized by growth and mental retardation [35][36][37]. Additionally, homozygous frameshift mutations in TRMT1, a writer for m2,2G, have been linked to intellectual disability [38]. Mutations and polymorphisms in FTSJ1, a writer for 2′O-methylribose, have also been linked to X-linked mental retardation [39][40][41][42][43]. Furthermore, targets of fragile X mental retardation protein (FMRP), a protein that is commonly mutated in Fragile X Syndrome, were enriched for m6A, and FMRP targets were targeted for degradation by YTHDF2 [44].
2.2.1. Alzheimer’s Disease
2.2.2. Major Depressive Disorder
2.3. Metabolic Disorders and Diseases
2.3.1. Obesity
2.3.2. Diabetes
-cell biology [63][64]. FTO mRNA expression was also higher in some T2DM patients [65]. Additionally, METTL3/METTL14 expression was decreased in β-cells of patients with T2DM, and METTL14 specifically may be essential for insulin secretion and β
2.3.3. Non-Alcoholic Fatty Liver Disease
Increased expression of fto was induced by a high-fat diet, resulting in increased lipogenesis and induction of non-alcoholic fatty liver disease (NAFLD), a disease that is commonly associated with obesity [68][69][70]. Identifying the m6A-dependent and m6A-independent functions of FTO in this context requires future study.
References
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T.K.; de Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–D307.
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42.
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485.
- Dai, Q.; Moshitch-Moshkovitz, S.; Han, D.; Kol, N.; Amariglio, N.; Rechavi, G.; Dominissini, D.; He, C. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat. Methods 2017, 14, 695–698.
- Dinescu, S.; Ignat, S.; Lazar, A.D.; Constantin, C.; Neagu, M.; Costache, M. Epitranscriptomic Signatures in lncRNAs and Their Possible Roles in Cancer. Genes (Basel) 2019, 10, 52.
- Esteve-Puig, R.; Bueno-Costa, A.; Esteller, M. Writers, readers and erasers of RNA modifications in cancer. Cancer Lett. 2020, 474, 127–137.
- Pandolfini, L.; Barbieri, I.; Bannister, A.J.; Hendrick, A.; Andrews, B.; Webster, N.; Murat, P.; Mach, P.; Brandi, R.; Robson, S.C.; et al. METTL1 Promotes let-7 MicroRNA Processing via m7G Methylation. Mol. Cell 2019, 74, 1278–1290.e1279.
- Romano, G.; Veneziano, D.; Nigita, G.; Nana-Sinkam, S.P. RNA Methylation in ncRNA: Classes, Detection, and Molecular Associations. Front. Genet. 2018, 9.
- Motorin, Y.; Helm, M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA 2011, 2, 611–631.
- Helm, M.; Motorin, Y. Detecting RNA modifications in the epitranscriptome: Predict and validate. Nat. Rev. Genet. 2017, 18, 275–291.
- Bohnsack, K.E.; Höbartner, C.; Bohnsack, M.T. Eukaryotic 5-methylcytosine (m⁵C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease. Genes (Basel) 2019, 10, 102.
- Uddin, M.B.; Wang, Z.; Yang, C. Dysregulations of Functional RNA Modifications in Cancer, Cancer Stemness and Cancer Therapeutics. Theranostics 2020, 10, 3164–3189.
- Barbieri, I.; Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 2020, 20, 303–322.
- Delaunay, S.; Frye, M. RNA modifications regulating cell fate in cancer. Nat. Cell Biol. 2019, 21, 552–559.
- Huang, H.; Weng, H.; Deng, X.; Chen, J. RNA Modifications in Cancer: Functions, Mechanisms, and Therapeutic Implications. Annu. Rev. Cancer Biol. 2020, 4, 221–240.
- Huo, F.-C.; Zhu, Z.-M.; Pei, D.-S. N6-methyladenosine (m6A) RNA modification in human cancer. Cell Prolif. 2020, 53, e12921.
- Ianniello, Z.; Paiardini, A.; Fatica, A. N6-Methyladenosine (m6A): A Promising New Molecular Target in Acute Myeloid Leukemia. Front. Oncol. 2019, 9, 1–11.
- Liu, L.; Wang, Y.; Wu, J.; Liu, J.; Qin, Z.; Fan, H. N(6)-Methyladenosine: A Potential Breakthrough for Human Cancer. Mol. Ther. Nucleic Acids 2020, 19, 804–813.
- Niu, Y.; Wan, A.; Lin, Z.; Lu, X.; Wan, G. N6-Methyladenosine modification: A novel pharmacological target for anti-cancer drug development. Acta Pharm. Sin. B 2018, 8, 833–843.
- Song, H.; Liu, D.; Dong, S.; Zeng, L.; Wu, Z.; Zhao, P.; Zhang, L.; Chen, Z.-S.; Zou, C. Epitranscriptomics and epiproteomics in cancer drug resistance: Therapeutic implications. Signal Transduct. Target. Ther. 2020, 5, 193.
- Wang, S.; Chai, P.; Jia, R.; Jia, R. Novel insights on m6A RNA methylation in tumorigenesis: A double-edged sword. Mol. Cancer 2018, 17, 101.
- Yang, G.; Sun, Z.; Zhang, N. Reshaping the role of m6A modification in cancer transcriptome: A review. Cancer Cell Int. 2020, 20, 353.
- Dong, Z.; Cui, H. The Emerging Roles of RNA Modifications in Glioblastoma. Cancers (Basel) 2020, 12, 736.
- Fazi, F.; Fatica, A. Interplay Between N6-Methyladenosine (m6A) and Non-coding RNAs in Cell Development and Cancer. Front. Cell Dev. Biol. 2019, 7.
- Widagdo, J.; Anggono, V. The m6A-epitranscriptomic signature in neurobiology: From neurodevelopment to brain plasticity. J. Neurochem. 2018, 147, 137–152.
- Zhang, M.; Zhai, Y.; Zhang, S.; Dai, X.; Li, Z. Roles of N6-Methyladenosine (m(6)A) in Stem Cell Fate Decisions and Early Embryonic Development in Mammals. Front. Cell Dev. Biol. 2020, 8, 782.
- Chokkalla, A.K.; Mehta, S.L.; Vemuganti, R. Epitranscriptomic regulation by m6A RNA methylation in brain development and diseases. J. Cereb. Blood Flow Metab. 2020, 40, 2331–2349.
- Engel, M.; Chen, A. The emerging role of mRNA methylation in normal and pathological behavior. Genes Brain Behav. 2018, 17, e12428.
- Ma, C.; Chang, M.; Lv, H.; Zhang, Z.-W.; Zhang, W.; He, X.; Wu, G.; Zhao, S.; Zhang, Y.; Wang, D.; et al. RNA m6A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 2018, 19, 68.
- Zhang, X.; Trebak, F.; Souza, L.A.C.; Shi, J.; Zhou, T.; Kehoe, P.G.; Chen, Q.; Feng Earley, Y. Small RNA modifications in Alzheimer’s disease. Neurobiol. Dis. 2020, 145, 105058.
- Geula, S.; Moshitch-Moshkovitz, S.; Dominissini, D.; Mansour, A.A.; Kol, N.; Salmon-Divon, M.; Hershkovitz, V.; Peer, E.; Mor, N.; Manor, Y.S.; et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 2015, 347, 1002–1006.
- Wang, C.X.; Cui, G.S.; Liu, X.; Xu, K.; Wang, M.; Zhang, X.X.; Jiang, L.Y.; Li, A.; Yang, Y.; Lai, W.Y.; et al. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 2018, 16, e2004880.
- Boissel, S.; Reish, O.; Proulx, K.; Kawagoe-Takaki, H.; Sedgwick, B.; Yeo, G.S.H.; Meyre, D.; Golzio, C.; Molinari, F.; Kadhom, N.; et al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am. J. Hum. Genet. 2009, 85, 106–111.
- Torres, A.G.; Batlle, E.; Ribas de Pouplana, L. Role of tRNA modifications in human diseases. Trends Mol. Med. 2014, 20, 306–314.
- Abbasi-Moheb, L.; Mertel, S.; Gonsior, M.; Nouri-Vahid, L.; Kahrizi, K.; Cirak, S.; Wieczorek, D.; Motazacker, M.M.; Esmaeeli-Nieh, S.; Cremer, K.; et al. Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2012, 90, 847–855.
- Blanco, S.; Dietmann, S.; Flores, J.V.; Hussain, S.; Kutter, C.; Humphreys, P.; Lukk, M.; Lombard, P.; Treps, L.; Popis, M.; et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014, 33, 2020–2039.
- Martinez, F.J.; Lee, J.H.; Lee, J.E.; Blanco, S.; Nickerson, E.; Gabriel, S.; Frye, M.; Al-Gazali, L.; Gleeson, J.G. Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J. Med. Genet. 2012, 49, 380–385.
- Najmabadi, H.; Hu, H.; Garshasbi, M.; Zemojtel, T.; Abedini, S.S.; Chen, W.; Hosseini, M.; Behjati, F.; Haas, S.; Jamali, P.; et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 2011, 478, 57–63.
- Bonnet, C.; Grégoire, M.J.; Brochet, K.; Raffo, E.; Leheup, B.; Jonveaux, P. Pure de-novo 5 Mb duplication at Xp11.22–p11.23 in a male: Phenotypic and molecular characterization. J. Hum. Genet. 2006, 51, 815.
- Dai, L.; Xing, L.; Gong, P.; Zhang, K.; Gao, X.; Zheng, Z.; Zhou, J.; Guo, Y.; Guo, S.; Zhang, F. Positive association of the FTSJ1 gene polymorphisms with nonsyndromic X-linked mental retardation in young Chinese male subjects. J. Hum. Genet. 2008, 53, 592–597.
- Freude, K.; Hoffmann, K.; Jensen, L.R.; Delatycki, M.B.; des Portes, V.; Moser, B.; Hamel, B.; van Bokhoven, H.; Moraine, C.; Fryns, J.P.; et al. Mutations in the FTSJ1 gene coding for a novel S-adenosylmethionine-binding protein cause nonsyndromic X-linked mental retardation. Am. J. Hum. Genet. 2004, 75, 305–309.
- Ramser, J.; Winnepenninckx, B.; Lenski, C.; Errijgers, V.; Platzer, M.; Schwartz, C.E.; Meindl, A.; Kooy, R.F. A splice site mutation in the methyltransferase gene FTSJ1 in Xp11.23 is associated with non-syndromic mental retardation in a large Belgian family (MRX9). J. Med. Genet. 2004, 41, 679–683.
- Takano, K.; Nakagawa, E.; Inoue, K.; Kamada, F.; Kure, S.; Goto, Y. A loss-of-function mutation in the FTSJ1 gene causes nonsyndromic X-linked mental retardation in a Japanese family. Am. J. Med. Genet. B Neuropsychiatry Genet. 2008, 147b, 479–484.
- Zhang, F.; Kang, Y.; Wang, M.; Li, Y.; Xu, T.; Yang, W.; Song, H.; Wu, H.; Shu, Q.; Jin, P. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 2018, 27, 3936–3950.
- Han, M.; Liu, Z.; Xu, Y.; Liu, X.; Wang, D.; Li, F.; Wang, Y.; Bi, J. Abnormality of m6A mRNA Methylation Is Involved in Alzheimer’s Disease. Front. Neurosci. 2020, 14.
- Reitz, C.; Tosto, G.; Mayeux, R.; Luchsinger, J.A. Genetic variants in the Fat and Obesity Associated (FTO) gene and risk of Alzheimer’s disease. PLoS ONE 2012, 7, e50354.
- Engel, M.; Eggert, C.; Kaplick, P.M.; Eder, M.; Röh, S.; Tietze, L.; Namendorf, C.; Arloth, J.; Weber, P.; Rex-Haffner, M.; et al. The Role of m(6)A/m-RNA Methylation in Stress Response Regulation. Neuron 2018, 99, 389–403.e389.
- Milaneschi, Y.; Lamers, F.; Mbarek, H.; Hottenga, J.J.; Boomsma, D.I.; Penninx, B.W.J.H. The effect of FTO rs9939609 on major depression differs across MDD subtypes. Mol. Psychiatry 2014, 19, 960–962.
- Samaan, Z.; Anand, S.; Zhang, X.; Desai, D.; Rivera, M.; Pare, G.; Thabane, L.; Xie, C.; Gerstein, H.; Engert, J.C.; et al. The protective effect of the obesity-associated rs9939609 A variant in fat mass- and obesity-associated gene on depression. Mol. Psychiatry 2013, 18, 1281–1286.
- Spychala, A.; Rüther, U. FTO affects hippocampal function by regulation of BDNF processing. PLoS ONE 2019, 14, e0211937.
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Zeggini, E.; Freathy, R.M.; Lindgren, C.M.; Perry, J.R.B.; Elliott, K.S.; Lango, H.; Rayner, N.W.; et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science (N. Y.) 2007, 316, 889–894.
- Andreasen, C.H.; Stender-Petersen, K.L.; Mogensen, M.S.; Torekov, S.S.; Wegner, L.; Andersen, G.; Nielsen, A.L.; Albrechtsen, A.; Borch-Johnsen, K.; Rasmussen, S.S.; et al. Low physical activity accentuates the effect of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes 2008, 57, 95–101.
- Scuteri, A.; Sanna, S.; Chen, W.M.; Uda, M.; Albai, G.; Strait, J.; Najjar, S.; Nagaraja, R.; Orrú, M.; Usala, G.; et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007, 3, e115.
- Cecil, J.E.; Tavendale, R.; Watt, P.; Hetherington, M.M.; Palmer, C.N. An obesity-associated FTO gene variant and increased energy intake in children. N. Engl. J. Med. 2008, 359, 2558–2566.
- Grant, S.F.; Li, M.; Bradfield, J.P.; Kim, C.E.; Annaiah, K.; Santa, E.; Glessner, J.T.; Casalunovo, T.; Frackelton, E.C.; Otieno, F.G.; et al. Association analysis of the FTO gene with obesity in children of Caucasian and African ancestry reveals a common tagging SNP. PLoS ONE 2008, 3, e1746.
- Hotta, K.; Nakata, Y.; Matsuo, T.; Kamohara, S.; Kotani, K.; Komatsu, R.; Itoh, N.; Mineo, I.; Wada, J.; Masuzaki, H.; et al. Variations in the FTO gene are associated with severe obesity in the Japanese. J. Hum. Genet. 2008, 53, 546–553.
- Prakash, J.; Srivastava, N.; Awasthi, S.; Agarwal, C.G.; Natu, S.M.; Rajpal, N.; Mittal, B. Association of FTO rs17817449 SNP with obesity and associated physiological parameters in a north Indian population. Ann. Hum. Biol. 2011, 38, 760–763.
- Qureshi, S.A.; Mumtaz, A.; Shahid, S.U.; Shabana, N.A. rs3751812, a common variant in fat mass and obesity-associated (FTO) gene, is associated with serum high- and low-density lipoprotein cholesterol in Pakistani individuals. Nutrition 2017, 39–40, 92–95.
- Fischer, J.; Koch, L.; Emmerling, C.; Vierkotten, J.; Peters, T.; Brüning, J.C.; Rüther, U. Inactivation of the Fto gene protects from obesity. Nature 2009, 458, 894–898.
- Church, C.; Moir, L.; McMurray, F.; Girard, C.; Banks, G.T.; Teboul, L.; Wells, S.; Brüning, J.C.; Nolan, P.M.; Ashcroft, F.M.; et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat. Genet. 2010, 42, 1086–1092.
- Zhao, X.; Yang, Y.; Sun, B.-F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.-J.; Ping, X.-L.; Chen, Y.-S.; Wang, W.-J.; et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–1419.
- Wu, J.; Frazier, K.; Zhang, J.; Gan, Z.; Wang, T.; Zhong, X. Emerging role of m6A RNA methylation in nutritional physiology and metabolism. Obes. Rev. 2020, 21, e12942.
- De Jesus, D.F.; Zhang, Z.; Kahraman, S.; Brown, N.K.; Chen, M.; Hu, J.; Gupta, M.K.; He, C.; Kulkarni, R.N. m6A mRNA methylation regulates human β-cell biology in physiological states and in type 2 diabetes. Nat. Metab. 2019, 1, 765–774.
- Wang, Y.; Sun, J.; Lin, Z.; Zhang, W.; Wang, S.; Wang, W.; Wang, Q.; Ning, G. m6A mRNA Methylation Controls Functional Maturation in Neonatal Murine β-Cells. Diabetes 2020, 69, 1708–1722.
- Shen, F.; Huang, W.; Huang, J.-T.; Xiong, J.; Yang, Y.; Wu, K.; Jia, G.-F.; Chen, J.; Feng, Y.-Q.; Yuan, B.-F.; et al. Decreased N6-Methyladenosine in Peripheral Blood RNA From Diabetic Patients Is Associated With FTO Expression Rather Than ALKBH5. J. Clin. Endocrinol. Metab. 2015, 100, E148–E154.
- Liu, J.; Luo, G.; Sun, J.; Men, L.; Ye, H.; He, C.; Ren, D. METTL14 is essential for β-cell survival and insulin secretion. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 2138–2148.
- Xie, W.; Ma, L.L.; Xu, Y.Q.; Wang, B.H.; Li, S.M. METTL3 inhibits hepatic insulin sensitivity via N6-methyladenosine modification of Fasn mRNA and promoting fatty acid metabolism. Biochem. Biophys. Res. Commun. 2019, 518, 120–126.
- Guo, J.; Ren, W.; Li, A.; Ding, Y.; Guo, W.; Su, D.; Hu, C.; Xu, K.; Chen, H.; Xu, X.; et al. Fat Mass and Obesity-Associated Gene Enhances Oxidative Stress and Lipogenesis in Nonalcoholic Fatty Liver Disease. Dig. Dis. Sci. 2013, 58, 1004–1009.
- Zhang, J.; Li, S.; Li, J.; Han, C.; Wang, Z.; Li, C.; Wang, X.; Liu, Z.; Wen, J.; Zheng, L. Expression and significance of fat mass and obesity associated gene and forkhead transcription factor O1 in non-alcoholic fatty liver disease. Chin. Med. J. (Engl.) 2014, 127, 3771–3776.
- Benedict, M.; Zhang, X. Non-alcoholic fatty liver disease: An expanded review. World J. Hepatol. 2017, 9, 715–732.





