RNA modifications are diverse post-transcriptional modifications that regulate RNA metabolism and gene expression. RNA modifications, and the writers, erasers, and readers that catalyze these modifications, serve as important signaling machineries in cellular stress responses and disease pathogenesis.
- RNA modifications
- disease
1. Introduction
RNA modifications are covalent chemical modifications of RNA molecules. To date, over 100 chemical modifications of RNA species have been identified [1][2][1,2]. The regulation and function of RNA modifications have recently emerged as pivotal mechanisms that regulate a wide range of biological and pathological processes, giving rise to the field known as epitranscriptomics.
RNA modifications are regulated through the coordination of ‘writers’, ‘erasers’ and ‘readers’, which deposit, remove, and recognize RNA modifications, respectively (Figure 1A). These enzymes represent key elements in patterning the epitranscriptomic landscape.
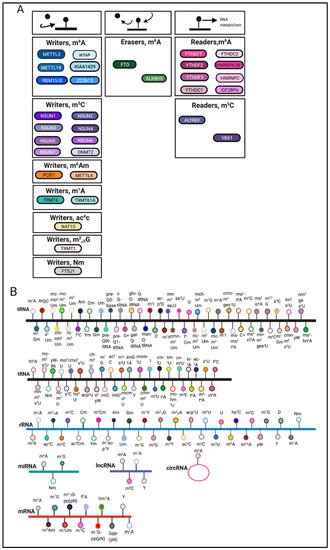
RNA modifications can occur on various RNA species including mRNA, tRNA, rRNA and other non-coding RNAs [9][3] (Figure 1B). Among these RNA species, transfer RNAs (tRNAs) contain the most RNA modifications [10][4]. One of the most abundant modifications on tRNA and rRNA is 5-methylcytosine (m5C) [11][5]. In comparison, mRNA modifications were more difficult to identify and characterize due to their low-abundance. The advent of sophisticated sequencing technologies and methods has generated a renewed interest in mRNA modifications.
2. RNA Modifications in Diseases
2.1. Cancer
The role of RNA modifications in cancer is cell-type and context-dependent and has been reviewed extensively in various types of cancer [12][13][14][15][16][17][18][19][20][21][22][16,72,73,74,75,76,77,78,79,80,81] and are highlighted in Figure 24. Rare RNA modifications have been described in cancer type-specific contexts, such as glioblastoma, and are detailed in [23][82]. An active area of research seeks to elucidate the role of RNA modifications in non-coding RNAs and other RNA species in the context of cancer, which are reviewed elsewhere [24][83].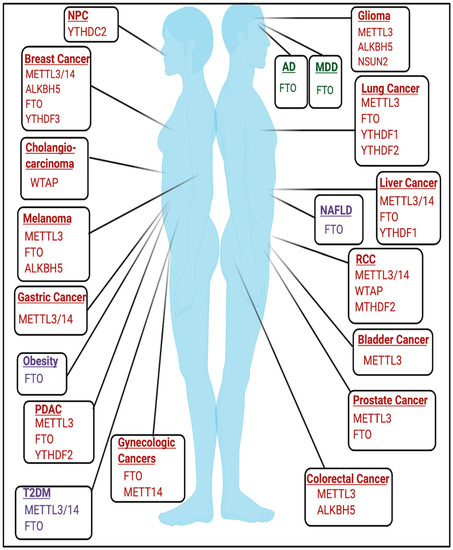
2.2. Developmental and Neurologic Disorders
The role of RNA modifications in the context of developmental and neurological disorders remains an active area of study. m6A has been previously found to play important roles in embryonic development and neurobiological functions [25][26][84,85]. The roles of m6A and other RNA modifications in mediating neurologic function are further discussed elsewhere [25][27][28][29][30][84,86,87,88,89].
The necessity of m6A in development is emphasized by early embryonic lethality in mettl3 KO mice [31][90]. Conditional mettl3 knockout in murine brains also resulted in severe developmental defects within the cerebrum and cortex and induced apoptosis in cerebella granule cells (CGCs) [32][91]. FTO may also be important in mediating development as expression of catalytically inactive mutant FTO(R316Q) resulted in severe growth defects [33][92]. Mutations in tRNA methyltransferases have been implicated in developmental disorders and are detailed in [34][93]. NSUN2 mutations have been linked to microcephaly, intellectual disability, and Dubowitz Syndrome, which is characterized by growth and mental retardation [35][36][37][94,95,96]. Additionally, homozygous frameshift mutations in TRMT1, a writer for m2,2G, have been linked to intellectual disability [38][97]. Mutations and polymorphisms in FTSJ1, a writer for 2′O-methylribose, have also been linked to X-linked mental retardation [39][40][41][42][43][98,99,100,101,102]. Furthermore, targets of fragile X mental retardation protein (FMRP), a protein that is commonly mutated in Fragile X Syndrome, were enriched for m6A, and FMRP targets were targeted for degradation by YTHDF2 [44][103].
2.2.1. Alzheimer’s Disease
2.2.2. Major Depressive Disorder
2.3. Metabolic Disorders and Diseases
FTO is a major driver in contributing to the pathogenesis of several metabolic diseases and therefore serves as a therapeutic target in this context.2.3.1. Obesity
2.3.2. Diabetes
-cell biology [63][64][122,123]. FTO mRNA expression was also higher in some T2DM patients [65][124]. Additionally, METTL3/METTL14 expression was decreased in β-cells of patients with T2DM, and METTL14 specifically may be essential for insulin secretion and β
-cell survival [63][64][66][122,123,125]. However, the role of m6A in mediating T2DM may be tissue and context-dependent as METTL3 and m6A levels were increased in liver tissue from T2DM patients [67][126].2.3.3. Non-Alcoholic Fatty Liver Disease
Increased expression of fto was induced by a high-fat diet, resulting in increased lipogenesis and induction of non-alcoholic fatty liver disease (NAFLD), a disease that is commonly associated with obesity [68][69][70][26,127,128]. Identifying the m6A-dependent and m6A-independent functions of FTO in this context requires future study.
