
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Maria Eugenia Moran-Diez | + 4991 word(s) | 4991 | 2021-04-26 08:38:32 | | | |
| 2 | Vivi Li | Meta information modification | 4991 | 2021-05-06 12:57:14 | | |
Video Upload Options
There is no doubt that Trichoderma is an inhabitant of the rhizosphere that plays an important role in how plants interact with the environment. Beyond the production of cell wall degrading enzymes and metabolites, Trichoderma spp. can protect plants by inducing faster and stronger immune responses, a mechanism known as priming, which involves enhanced accumulation of dormant cellular proteins that function in intracellular signal amplification. One example of these proteins is the mitogen-activated protein kinases (MAPK) that are triggered by the rise of cytosolic calcium levels and cellular redox changes following a stressful challenge. Transcription factors such as WRKYs, MYBs, and MYCs, play important roles in priming as they act as regulatory nodes in the transcriptional network of systemic defence after stress recognition.
1. Introduction
Trichoderma is one of the most studied genera of ascomycetous fungi because of the importance of the practical applications and particular skills of the species that compose it [1][2]. Possibly, its main characteristic is the ability to exert positive effects on plants by means of the production of cell wall degrading enzymes (CWDE) [3] and metabolites with antimicrobial activity [4], and the release of volatile organic compounds (VOC) [5][6], which can act together as direct biocontrol agents on phytopathogenic fungi, oomycetes, nematodes, and bacteria [7]. Trichoderma spp. also compete with other plant beneficial microbes and impact the arbuscular mycorrhizal fungi (AMF) abundance in bulk soil microbiomes in agricultural systems [8]. In this sense, a single application of T. harzianum increased the rhizosphere levels and facilitated the access of AMF to the roots of the host (e.g., wheat) [8] and non-host (e.g., rapeseed, Arabidopsis) [9] plants. Trichoderma spp. can also act as indirect biocontrol agents by activating systemic immune responses in a coordinated way [10], resulting in faster and stronger induction of plant basal resistance mechanisms upon the perception of a later triggering stimulus. This phenomenon is known as priming of defence [11] and Trichoderma can provide the plant with long-lasting resistance against biotic and abiotic stresses by balancing the different phytohormone-dependent pathways—among which salicylic acid (SA), jasmonates (JA), ethylene (ET), abscisic acid (ABA), auxin (indole-3-acetic acid: IAA), and gibberellins (GA) are the most relevant—and modulating the levels of growth and defence regulatory proteins [12]. The advantage to the plant being primed for particular stress responses is in facilitating a faster and stronger reaction if the stress recurs [13]. Reinforced responses to pathogen attacks come under the category of induced defence, while responses to abiotic stresses are referred to as acclimation or hardening, even though these responses are very similar in their genesis and can also be enhanced by priming treatments [13]. An accurate definition of how Trichoderma exerts its beneficial action on plants is of particular relevance to the way in which commercial products based on the abilities of Trichoderma are registered.
Regardless of whether Trichoderma is recognized by the plant through cell-surface pattern recognition receptors (PRRs), priming involves multiple cellular localizations of targets such as: (i) inactive metabolites stored in vacuoles, which can be hydrolysed and released into the cytoplasm; (ii) reactive oxygen species (ROS) that can also interact with phytohormone-dependent pathways in the cytoplasm; and (iii) epigenetic changes that occur in the nucleus [14]. Cascades of mitogen-activated protein kinases (MAPK) and Ca2+-dependent protein kinases (CDPK, especially related to plant growth regulation and acclimation, [15]) function downstream of the ligand binding and the subsequent PRR activation and transmit extracellular priming stimuli into intracellular responses, while amplifying at the same time the transduction signals to the nucleus, resulting in transcriptional reprogramming, callose deposition, and, ultimately, synthesis of hormones and activation of defence-related genes [16].
Priming involves enhanced accumulation of dormant cellular proteins functioning in intracellular signal amplification [11][17], as is the case with: (i) signalling proteins in an inactive configuration that are activated upon exposure to stress, as occurs with MAPKs being triggered by the rise of cytosolic Ca2+ levels and cellular redox changes following a stressful challenge; (ii) transcription factors (TF) (e.g., auxin response factors (ARF), ethylene response factors (ERF), MYBs, MYCs, WRKYs) acting as regulatory nodes in the transcriptional network of systemic defence after stress recognition; and (iii) epigenetic regulation involving histone modifications and replacements, DNA (de)methylation, and RNA-directed DNA methylation (RdDM) [13][14][16][17][18][19][20].
In plant–Trichoderma systems, the actions of one trigger counteract the other. We refer to a dynamic process, which follows its own zig-zag defence model [1], in which four timing stages might be identified—Stage 1: the plant’s early perception of Trichoderma that only lasts a few hours; Stage 2: cell signalling, giving rise to systemic defence responses that can reach several days, gradually decaying in weeks; Stage 3: long-lasting priming; Stage 4: transgenerational inheritance. The latter two stages last from a few weeks to the offspring stage. Here, based on our own and other authors’ experimental evidence, we want to present how plants respond to the stimuli caused by Trichoderma, stressing that such responses are transmitted onto the progeny, in terms of different types of defence, acclimation, and growth control.
2. Plant’s Early Perception of Trichoderma
It has been observed that the root endospheric microbial community is less diverse than that of the rhizosphere, which indicates that many fungi in the rhizosphere might be trying to enter the plant, but only a few among them are allowed and prove successful [8][21]. This is the case for Trichoderma that manages to use the needed mechanisms in order to achieve its aims. Trichoderma activates PRRs by means of microbe- or damage-associated molecular patterns (MAMP or DAMP), apoplastic effectors, and a variety of VOCs [12][22][23][24]. Then, oxidative burst, callose deposition, and Ca2+ and ROS signalling are initiated rapidly following the attack because, at that point in its perception of Trichoderma, the plant does not recognize that it is a friendly attack. Secreted Trichoderma molecules are sensed by plant cells through intracellular Ca2+ changes that lead to not only intracellular ROS accumulation but to the necessary adjustments to prime defence mechanisms [25]. Figure 1 summarizes some of the main effects and responses triggered during the early interaction between the plant and Trichoderma. It is well established that G-proteins located within the cell membrane can be the linkage between host-derived signals and Trichoderma-intracellular signalling pathways, resulting in an increase in mycoparasitism-relevant processes such as the production of fungal CWDEs and antifungal metabolites, and the formation of infection structures [26]. Although the activation of G-protein signalling is different in animals and fungi than in plants, it can be thought that G-proteins may play a role in the recognition of Trichoderma by the plant. In this way, heterotrimeric G-proteins function as a convergence point of plant defence signalling by mediating responses initiated by the activation of different PRRs, as is the case of systemically transmitted stomatal closure, ROS production, or callose deposition at the cell wall [27]. There is also evidence for the role of G-protein-dependent regulation downstream of PRRs in plant growth and development [28], which indicates a molecular mechanism of G-protein recognition and signalling activation/deactivation compatible with the effects that Trichoderma has on plants. This fact constitutes a promising line of research to understand the initial dialogue between Trichoderma and the plant.
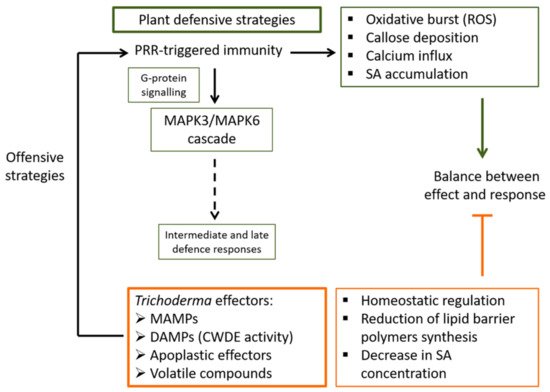
Figure 1. Schematic outline of plant’s early response to the interaction with Trichoderma. Plant´s cell surface pattern recognition receptors (PRR) are the first line of defence against Trichoderma effectors during the early response (this first stage of the colonization being considered by the plant as an attack). As a result of this first encounter between the plant and Trichoderma, a series of events will be triggered in the plant including the rapid release of reactive oxygen species (ROS), the deposition of callose, calcium influx, and accumulation of salicylic acid (SA) at the primary inoculation site as well as for systemic defence. Another one of the early events triggered by PRR stimulation is the activation of the mitogen-activated protein kinases (MAPK) cascade mediated by the G-protein complex, which will lead to several intermediate and late defence responses. Trichoderma spp. will find their way to balance this early plant response into one of mutual benefit by regulating homeostasis and reducing the synthesis of plant lipid barrier polymers, as well as by decreasing the production of SA.
Most of the work on Trichoderma–plant interactions has been carried out in root systems. However, it should not be forgotten that many Trichoderma strains live and can be isolated from the aboveground parts of the plant [29], where they endophytically colonize leaves and stems [30]. Some strains of Trichoderma are effective in the direct control of pathogens in the phyllosphere, although their mechanism of action seems to be linked more to induced resistance and mycoparasitism than to direct competition [31]. Selected Trichoderma strains may also release VOCs related to plant growth promotion [32] but may also be involved in reducing energy losses and inducing photosynthesis and resistance to foliar pathogens, such as those producing powdery mildew [33]. Even endophytic Trichoderma strains can mitigate the negative impact of leaf-cutting ants in both agriculture and silviculture due to the potential to act as bodyguards to their plant hosts affording protection to the whole plant [34].
ROS production is one of the earliest responses, starting only a few minutes after the PRRs switch on, although, it also occurs later when the defence is already established, but at a much slower pace. ROS have been proposed to act as antimicrobials, as cross-linkers of the plant cell wall to block pathogen entry, and as local and systemic secondary messengers to trigger additional immune and developmental responses [35]. When colonizing the roots, Trichoderma spp. go beyond the oxidative burst generated as an early defence response by the plant. So, Trichoderma strains take advantage of their ability to compete in the rhizosphere, since they have the particularity of tolerating high levels of ROS [36] and even produce ROS by themselves, which in return facilitates the biocontrol activity against pathogens, most of them with a lower range of oxidative stress tolerance [37]. The establishment of Trichoderma in the root tissues may be fostered by the action of plant CWDEs such as hemicellulases and polygalacturonases. In T. harzianum, endopolygalacturonase activity was required for active root colonization and the release of oligogalacturonides, oligomers of α-1,4-linked galacturonic acid generated by the partial hydrolysis of pectin, capable of acting as DAMPs to activate the systemic defences against the necrotrophic leaf pathogenic fungus Botrytis cinerea [38]. During the colonization of the root, SA is the key phytohormone to reinforce plant cell walls, restricting Trichoderma to the apoplast and preventing the arrival to the vascular system. The important role of SA in Trichoderma root colonization was demonstrated with Arabidopsis sid2 mutants unable to synthesize SA, as plants lacking this phytohormone did not accumulate callose and allowed the dissemination of Trichoderma in the vascular ring and the upward movement to the aboveground part of the plant, which would lead to a subsequent collapse [39]. Therefore, it seems clear that Trichoderma has to cope with the oxidative burst, callose deposition, and SA-signalized immunity during the first hours of the interaction, when the early SA-dependent defence response may not reach its full potential. This struggle allows Trichoderma to colonize the root, although the down-regulation of SA would reach a balanced level where there is no indiscriminate spread of the fungus [22]. Thus, one of the first tasks Trichoderma needs to do is to accurately overcome and counteract the plant’s initial defences. In this regard, we have observed a strong down-regulation of the SA-responsive PATHOGENESIS-RELATED (PR) PROTEIN 1 (PR-1) gene after 6 h of cultivating Arabidopsis in the presence of T. harzianum [40], and it has also been reported as early as after 4 h of Arabidopsis–Trichoderma interaction, as a strong down-regulation of the SA-related ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) gene. EDS1 operates upstream of SA-dependent defences and is required early to accumulate SA, although it is also required later to generate the plant hypersensitive response (HR) to prevent the spread of infection by microbial pathogens [41]. SA production is not the only defence mechanism limited early on by the effect of Trichoderma. Syntheses of lipid barrier polymers and signalling during pathogen attack [42] are down-regulated in Arabidopsis root tissues 24 h post inoculation (hpi) with T. harzianum [40]. In this way, the rapid and local silencing of early plant defence response enables Trichoderma to establish symbiotic interactions, as it has been observed for other symbiotic systems [43]. The down-regulation of PR proteins in maize seedling roots treated with T. afroharzianum is in agreement with the local silencing of defence genes to allow fungal growth into the roots [44]. However, following the decrease of defences in Arabidopsis root colonized by Trichoderma, the expression of both PR-1 and EDS1 genes was increased as early as 48 h [40]. In addition, the accumulation of PR proteins and catalase (CAT) activity increased in both the roots and leaves of cucumber seedlings at 48 hpi with T. asperelloides T203 [45]. Corroborating this fact, it has also been described that tomato plants showed a sharped increase in the expression of the PR-1b1 gene at 72 hpi with T. longibrachiatum [46].
Based on the assumptions that each plant–Trichoderma interaction is very particular, that different activations of defence-related genes occur among Trichoderma strains against the same pathogen [47], and that it is difficult to establish a specific time point when early defence responses end, it has been proposed that 48 hpi would indicate the moment of transition when the plant reprograms its transcriptional machinery mainly towards redox and defence processes, fully accepting that Trichoderma is not an enemy [48]. There is no doubt that Trichoderma needs to minimize levels of SA-dependent defence responses—or at least prevent them from rising—but the maintenance of certain levels of SA is also very important in the establishment of an early oxidative burst. ROS generation, including superoxide anions and H2O2, is indicative of early defence response activation since they do not only act directly as toxic agents against the host plant cells but also against the pathogens by killing them or ceasing their virulent activity [49]. SA increases the accumulation of H2O2 in plants after pathogen infection. However, as observed in the first 24–48 h of tomato–T. erinaceum interaction, plants challenged with Trichoderma accumulated H2O2 at lower levels than those detected in plants infected with the pathogen Fusarium oxysporum f. sp. lycopersici (FOL) [50]. Similarly, a marked reduction in the H2O2 levels during the first 3–24 h of interaction between tomato plants and T. asperellum allowed the explanation of the protective effects exerted by Trichoderma against F. oxysporum and B. cinerea HR-like lesions [51]. These two cases may serve to illustrate that the plant accepts Trichoderma better than the pathogen.
Nevertheless, SA also plays opposite roles in preventing the damage to plants against oxidative stress by inducing the production of detoxifying enzymes such as CAT, superoxide dismutase (SOD), ascorbate peroxidase (APx), or glutathione S-transferase (GST), which reduce the accumulation of ROS as these molecules are not positive for plant cells either. We have observed that a GST gene strongly induced by SA, which participates in ROS detoxification, reinforcement of cell walls, formation of phytoalexins, and degradation of various toxic substances [52], was down-regulated in Arabidopsis plants at 24 hpi with T. harzianum [40]. In this sense, the treatment of Arabidopsis roots with T. harzianum activated, in just 4 h, the expression of the CYP71A12 gene encoding a cytochrome P450 required in the biosynthesis of phytoalexins, reached the maximum peak of expression at 24 hpi but experienced a subsequent drop after that point [53]. In the tomato–T. erinaceum study, the CAT and SOD enzymatic activities were higher in primed plants than in those unprimed with Trichoderma under FOL challenged conditions [50]. In addition, SOD rose from 24 to 48 h with a drop to 72 h, while CAT dropped to 48 h and rose sharply to 72 h. A maize–T. guizhouense study [54] found that H2O2 was accumulated in maize tissues at 24 hpi in the interaction with Trichoderma. Additionally, the expression of the auxin synthesis related gene YUC4, was down-regulated at 12 hpi and at 48 hpi, with a peak of expression at 24 hpi, which indicates a decline in SA-dependent defences and lateral root growth. The examples of the undulating behaviour of plant responses given here can be explained by SOD gene up-regulation resulting in diminished ROS activity and therefore higher accumulation of H2O2, which in turn, induces subsequent CAT activity. After the initial oxidative burst, plant performance would be justified because ROS also functions at longer times as signalling regulators of growth and mitigation of biotic and abiotic stresses. If to all this we add that Trichoderma produces its own ROS to compete with other inhabitants of the rhizosphere [37], it would seem clear that Trichoderma works by fine-tuning the levels of ROS in the plant.
3. Systemic Plant Responses to Trichoderma
Stage 2 does not have a fixed starting time but depending on the system under study, it is considered not to start after 48–72 h of Trichoderma being applied. Upon root colonization, the plant produces a phytohormone blend (as a result of a series of transcription cascades that mainly involve SA, JA, and ET but also ABA, IAA, and GA signalling pathways) that constitutes the Trichoderma signal signature [10]. The signalling pathways that are activated by the endogenous accumulation of these phytohormone signals not only regulate the plant’s dialogue with Trichoderma but also contribute to deploying different active defence responses that are effective against distinct classes of attackers [55]. The effectiveness of the contribution of Trichoderma to protecting plants will depend on how the pathogens may have evolved to manipulate plants for their own benefit, interfering with the plant’s defence-signalling network [47]. At 72 hpi, changes in SA-induced defences may contribute to hindering further Trichoderma penetration but may also be indicating induced priming against other attackers [48]. Once the colonization has begun, the Trichoderma growth inside roots is restricted to the intercellular spaces of the cortex and epidermis, because callose deposition prevents access to the vascular bundles [45]. The endophytic accumulation of Trichoderma will also act as a physical barrier to prevent the movement of the pathogen in the plant [56]. Trichoderma strains from different species can colonize the rhizosphere and grow within the root tissue not only without causing damage but while priming plants against biotic and abiotic stresses [2].
It has been observed that upon challenge exposure to stress stimuli, elevated accumulation of mRNA transcripts and inactive MAPK proteins, such as MPK3, primes Arabidopsis plants for enhanced defence and provokes immunity to future invasions, whereas their down-regulation reduces immunity [57]. In this sense, the plant defence mediated by the activation of kinases MPK3 and MPK6 rapidly alters the expression of photosynthesis-related genes and photosynthetic activity, which promotes the accumulation of ROS and accelerates HR cell death [58]. The dual functionality of the MPK3/6 cascade in promoting defence and inhibiting photosynthesis potentially allows it to orchestrate the trade-off between plant growth and defence in plant immunity. Interestingly, Agrobacterium tumefaciens hijacks the MPK3/6 pathway for delivering the T-DNA into the plant cell nucleus [59]. MPK4 is another regulatory hub that is essential for basal resistance to invasive biotrophic and hemibiotrophic attackers that has a role in ROS signalling and negatively regulates the positive effects of EDS1 on SA-mediated defence, while also displaying positive regulation of JA/ET-dependent immunity [60].
In plant–Trichoderma interactions, root colonization was accompanied by the systemic expression of an MPK3 orthologous gene in cucumber plants challenged with T. asperelloides and the overexpression of MPK3 primed them for JA- and ET-mediated defences against P. syringae pv. lachrymans without previous root colonization by the beneficial fungus [61]. It was found in Arabidopsis that plant growth regulators, such as ET and IAA, produced by the plant in response to T. atroviride, control several aspects of the root growth but, simultaneously, they can modulate MPK6 activity affecting root development [62]. MPK6 also activates ET biosynthesis through the phosphorylation of 1-AMINOCYCLOPROPANE-1-CARBOXYLATE (ACC) synthase after PRR stimulation [63]. It was observed that after T. viride inoculation onto both sides of Arabidopsis leaves, MPK6 increased the expression of a plasma membrane proton ATPase, which had a regulatory role in further leaf colonization and plant growth promotion [32]. The MPK3/6 cascade regulates phytoalexin biosynthesis and the phosphorylation of the TF ERF6, triggering the ET signalling-dependent defences [64]. The MPK3/6 cascade leads to the activation of WRKY type TFs, such as WRKY33, culminating in the expression of defence genes. In this way, early transcriptional responses mediated by WRKY33 result in the down-regulation of SA-related plant defences, but also in the up-regulation of JA-associated responses at later stages [65], as reported in bean plants colonized with T. velutinum [66]. A summary of the TFs that have been reported to be relevant to the Trichoderma–plant interaction and their regulatory effect are presented in Table 1. Studies performed on Arabidopsis showed that bacterial infection resulted in increased accumulation of the metabolite azelaic acid (AZA) that has been shown to prime plants to accumulate higher levels of SA and the activation of systemic defence against the pathogen P. syringae [67]. AZA induces the expression of the AZELAIC ACID INDUCED 1 (AZI1) gene, which encodes the secreted lipid transfer protein AZI1, involved in the production and/or translocation of a mobile immune signal [68], and has proven to be one of the targets of MPK3 and MPK6 [69]. All these cases may be indicative that signal transduction by the MPK3/6 cascade is not only necessary to induce defence responses but also constitutes a molecular node that fine-tunes the phytohormone networking that gives rise to a balance between growth and defence. The quick activation of MPK4, that negatively regulates SA accumulation and which later on regulates the inhibitory crosstalk between the SA and JA/ET signalling networks, is in agreement with the rapid (4 h) down-regulation of EDS1 in tomato plants and the subsequent up-regulation of this gene at 48 hpi with T. harzianum [40]. The already indicated undulating responses of plants undoubtedly condition the goal of building up an effective systemic defence in the medium and long term.
In addition to the role of MPKs in the activation of WRKY33 within the nucleus, there are several examples where Trichoderma spp. are described as priming activators by modulating the expression of different TFs involved in transcriptional activation or repression at the end of the MAPK signal transduction cascades, resulting in accelerated activation of the defence responses followed by their subsequent moderation, which gives rise to the timing stages that lead to the different types of systemic resistance to stresses [47][70][71][72][73][74][75]. This Trichoderma trait is corroborated by the fact that cis-elements, known to be involved in stress responses, such as WRKY, MYB, and MYC motifs, have been found in promoters of secreted Trichoderma effectors [76][77].
TFs with highly conserved WRKY domains are quite common in plants (e.g., 38 genes in the moss Physcomitrella, 83 in tomato, and 197 in soybean) and play roles in the regulation of transcriptional reprogramming associated with plant stress responses against both biotic and abiotic factors. Genes induced during defence responses often contain WRKY TF-binding sites since WRKYs can interact (in)directly with MAMPs/DAMPs or effector proteins to activate or repress plant defences. In addition to the WRKY33 seen above being associated, respectively, with lowering and raising SA- and JA-dependent defences, experimental data pointed out the existence of crosstalk between induced defence and acclimation responses mediated by WRKYs [78]. The ankyrin repeat protein NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1) is a transcription cofactor that acts as the master key to the plant defence signalling network and mediates crosstalk between the SA and JA/ET responses [68].
WRKY54 and WRKY70 are direct transcriptional targets of NPR1 to negatively control SA accumulation [79]. WRKY54 and WRKY70 play dual roles in systemic defence, both as negative regulators of SA biosynthesis and as positive regulators of SA signalling. In a 24-h Arabidopsis root interaction with T. harzianum [40] and T. asperelloides [72], WRKY54 was down-regulated. WRKY54 and WRKY70 were also down-regulated in Arabidopsis plants at 24 and 48 hpi with T. atroviride, with slight gene expression increases from 48 h until the relationship between Arabidopsis and Trichoderma was stably established after six days [73]. These results confirmed the important role of NPR1 as a negative regulator of SA biosynthesis in plants challenged with Trichoderma but also as an SA signal transducer at longer times. The structurally related WRKY18, WRKY40, and WRKY60 get accumulated in response to SA and JA during biotic stress as well as to ABA during abiotic stress responses. Interestingly, these TFs have leucine zipper motifs at their N termini via which they directly bind to their own respective promoters, in order to negatively regulate their expression patterns to maintain the homeostasis of WRKY proteins, which implies an additional element to understanding the crosstalk between Trichoderma and plants. WRKY18, WRKY40, and WRKY60 are also activators of T-DNA genes after their integration in the plant genome following A. tumefaciens infection [80], which also links them to the regulatory role of MPK3/6 in balancing defence and growth and opens a debate on how the use of T-DNA genes can affect the regulatory systems of plants transformed with the Ti plasmid. In Arabidopsis plants treated with T. asperelloides, the expression of WRKY18 and WRKY40 was enhanced at 9 hpi, which correlated with the negative regulation of the SA-dependent defence positive regulators FLAVIN MONOOXYGENASE 1 (FMO1) and EDS1 at the same time point. The activation of JA-signalling genes via the suppression of JASMONATE ZIM DOMAIN (JAZ) repressors was also observed [72]. The progressive decline in the expression levels of WRKY18 and WRKY40 reinforces the undulating behaviour of the defence responses in Trichoderma-colonized plants. To cite just one example of this pattern, WRKY60 kept expression levels low in the first hours of plant–Trichoderma interaction and raised them after six days [72][73]. WRKY4 gene expression is induced by SA and pathogen infection, and functions as a suppressor of SA-dependent defence genes. It was down-regulated in the first 24–48 h of inoculation of T. erinaceum in tomato [50] and T. asperelloides in Arabidopsis [72], although the levels of expression of WRKY4 tended to increase over time. WRKY activation is not exclusive to MAPK cascades as it has been proven that CDPKs phosphorylate WRKY8 for immune gene expression and ROS production [81]. WRKY8 in fact was strongly up-regulated at 24 h of Arabidopsis-T. atroviride interaction and presented a gradual decrease in its expression from then on [73]. A special point of attention is the case of WRKY70, which is a primary target for the Arabidopsis histone methylase ARABIDOPSIS TRITHORAX-LIKE FACTOR 1 (ATX1) involved in establishing the trimethylation pattern of lysine 4 (K4) in histone H3 (H3K4me3), while the SA-responsive gene PR-1 and the JA-responsive gene THI2.1 are secondary targets. The finding that the PR-1 and THI2.1 nucleosomes carry H3K4me3-marks unrelated to their transcription states has suggested that SA- and JA-inducible defence genes keep their nucleosomes in an ‘active memory’ state in preparation for quick-changes of transcription when needed by the cell [82]. As we will see later, after an initial stimulus, chromatin remodelling is stored in the defence gene promoters, which can be activated when plants are subjected to new stresses [83]. Similarly, histone methylations at the WRKY40 promoter activate the SA-dependent pathway and negatively regulate ABA-dependent adaptation of plants to unfavourable environmental conditions [84]. This behaviour would be in accordance with what has been reported for Arabidopsis-T. asperelloides [72] and with the proposed plant–Trichoderma interaction model [10].
The R2R3-MYB TFs that only exist in terrestrial plants are capable of binding to the WRKY promoters leading to their expression (e.g., MYB44 directly regulates the expression of WRKY70; and MYB51 that of WRKY33) [85][86]. MYBs are involved in multiple stress responses, including defence-induced lignification and basal immunity (MYB15), SA-and JA-mediated defence responses and enhancement of ABA signalling (MYB44), indolic glucosinolate biosynthesis upon SA and ET signalling in Arabidopsis and salt/osmotic stress regulation (MYB51), early signalling of rhizosphere-induced systemic defence (MYB72), and auxin modulation and root development through interaction with ARFs (MYB77) [85][86][87][88][89]. The root-specific TF MYB72 is required for JA/ET-dependent systemic resistance and callose accumulation induced by T. asperellum in Arabidopsis plants and also functions as an early node of convergence in the signalling pathways that are induced by different beneficial microorganisms, such as Trichoderma itself and Pseudomonas fluorescens [70]. In Arabidopsis roots colonized by T. asperelloides [72], an undulating expression pattern was observed in different MYB genes, with maximum levels for MYB15, MYB51, and MYB72 at 24 h. At this time point, the growth-regulating MYB77 gene showed its minimal expression, limiting lateral root development while increasing defences, which would be corroborating the ability of Trichoderma to balance the costs of plant growth and defence [12].
BASIC HELIX-LOOP-HELIX (bHLH) MYC TFs regulate multiple functions in plants. In particular, MYC2 is required for JA-dependent systemic defences triggered by beneficial soil microbes, JA-regulated plant development, and lateral root formation. MYC2 acts as a regulatory hub within the JA signalling pathway and functions as a negative regulator of the action of MYB51 [90], cooperating also with MYB2 in the activation of the ABA signalling pathway in response to salt stress and water deficit [91]. MYC2 contains a JAZ interaction domain and a transcriptional activation domain which recruits the mediator of RNA polymerase II (Pol II) transcription subunit 25 (MED25) of the Mediator complex [90][92]. When the levels of JA are low, MYC2 binds JAZ proteins, leading to the repression of MYC2 activity, resulting in the suppression of JA-responsive gene expression. However, when JA concentration is elevated, JAZ repressors are degraded in the 26S proteasome and MYC2 is free to form a complex with MED25 to trigger the initiation and amplification of JA-mediated transcriptional responses [93]. Arabidopsis roots inoculated with T. harzianum showed up-regulation of MYB51 and the PROLINE RICH PROTEIN BstNI SUBFAMILY 1 (PRB1) gene involved in JA- and ET-dependent defence responses in root tissues in 4 h, and down-regulation of their expression over time, while the expression of the TF ERF significantly increased from 4 to 48 hpi compared to uninoculated plants [53]. This would indicate that: (i) in the first hours of interaction with Trichoderma, JA levels are high, in an attempt to counteract the activation of SA-dependent defences, and MYB51 is not being negatively regulated by MYC2; and (ii) ERF4 acts as a transcriptional repressor of the SA-mediated suppression of JA-responsive genes, as SA levels would be being restored while maintaining a JA-dependent defence [94].
Via JA signalling, MYC2 also coordinates defence- and growth-related processes. In fact, in plant–Trichoderma interactions, MYC2 has been revealed as a master regulator of balancing the costs of plant growth and defence, since it also regulates crosstalk between the JA-dependent pathways and those of other phytohormones [12][90][95]. Of particular interest throughout these processes are, on the side of MYC2, the MED25 binding domain required for relaying JA transducing signals to the Pol II transcriptional machinery and on the MED25 side, the activator-interacting domain essential for the binding with MYC and ERF TFs [92]. The MYC2-MED25 functional complex activates JA-dependent defences but also regulates the termination of JA signalling, which means that the expression of genes dependent on the JA defence acquires an undulating shape at certain time periods [96]. MED25 has been also described as a regulator of abiotic stress responses, interacting with promoter regions of ABA-INSENSITIVE5 (ABI5) TF, leading to a negative effect on ABA-regulated gene transcription [97], in the same way as histone methylation does in WRKY40 [78]. MED25 functions as a negative regulator of primary and lateral root growth through auxin-related mechanisms [98]. MED25 cooperates with other subunits of the Mediator, as is the case with: (i) MED16, which governs the JA-ET crosstalk that also binds WRKY33 and regulates the expression of JA responsive genes and the ERF-domain TF ORA59; and (ii) MED18, which regulates JA-SA crosstalk and ROS production [92]. Although MED25 and MYC2 are evolutionarily conserved, their relatively weak interaction is species-specific and may explain why they are not interchangeable among plant species [99]. So, the different MYC2 and MED25 binding abilities could help explain a part of the enormous capacity of plants to alternate and progressively modify their defences to adapt to a changing environment.
References
- Lorito, M.; Woo, S.L.; Harman, G.E.; Monte, E. Translational research on Trichoderma: From ’omics to the field. Annu. Rev. Phytopathol. 2010, 48, 395–417.
- Druzhinina, I.S.; Seidl-Seiboth, V.; Herrera-Estrella, A.; Horwitz, B.A.; Kenerley, C.M.; Monte, E.; Mukherjee, P.K.; Zeilinger, S.; Grigoriev, I.V.; Kubicek, C.P. Trichoderma: The genomics of opportunistic success. Nat. Rev. Microbiol. 2011, 9, 749–759.
- Sanz, L.; Montero, M.; Grondona, I.; Vizcaíno, J.A.; Llobell, A.; Hermosa, R.; Monte, E. Cell wall-degrading isoenzyme profiles of Trichoderma biocontrol strains show correlation with rDNA taxonomic species. Curr. Genet. 2004, 46, 277–286.
- Vizcaíno, J.A.; Sanz, L.; Basilio, A.; Vicente, F.; Gutiérrez, S.; Hermosa, M.R.; Monte, E. Screening of antimicrobial activities in Trichoderma isolates representing three Trichoderma sections. Mycol. Res. 2005, 109, 1397–1406.
- Keszler, Á.; Forgács, E.; Kótai, L.; Vizcaíno, J.A.; Monte, E.; García-Acha, I. Separation and identification of volatile components in the fermentation broth of Trichoderma atroviride by solid-phase extraction and gas chromatography-mass spectrometry. J. Chromatogr. Sci. 2000, 38, 421–424.
- Vinale, F.; Sivasithamparam, K.; Ghisalberti, E.L.; Marra, R.; Woo, S.L.; Lorito, M. Trichoderma-plant-pathogen interactions. Soil Biol. Biochem. 2008, 40, 1–10.
- Monte, E.; Hermosa, R. Deciphering biocontrol qualities and other plant beneficial traits in Trichoderma. In How Research Can Stimulate the Development of Commercial Biological Control Against Plant Diseases. Progress in Biological Control, 1st ed.; De Cal, A., Melgarejo, P., Magan, N., Eds.; Springer: Cham, Switzerland, 2020; Volume 21, pp. 95–108.
- Illescas, M.; Rubio, M.B.; Hernández-Ruiz, V.; Morán-Diez, M.E.; Martínez de Alba, A.E.; Nicolás, C.; Monte, E.; Hermosa, R. Effect of inorganic N top dressing and Trichoderma harzianum seed-inoculation on crop yield and the shaping of root microbial communities of wheat plants cultivated under high basal N fertilization. Front. Plant Sci. 2020, 11, 575861.
- Poveda, J.; Hermosa, R.; Monte, E.; Nicolás, C. Trichoderma harzianum favours the access of arbuscular mycorrhizal fungi to non-host Brassicaceae roots and increases plant productivity. Sci. Rep. 2019, 9, 1–11.
- Hermosa, R.; Viterbo, A.; Chet, I.; Monte, E. Plant-beneficial effects of Trichoderma and of its genes. Microbiology 2012, 158, 17–25.
- Conrath, U.; Beckers, G.J.M.; Langenbach, C.J.G.; Jaskiewicz, M.R. Priming for enhanced defense. Annu. Rev. Phytopathol. 2015, 53, 97–119.
- Hermosa, R.; Rubio, M.B.; Cardoza, R.E.; Nicolás, C.; Monte, E.; Gutiérrez, S. The contribution of Trichoderma to balancing the costs of plant growth and defense. Int. Microbiol. 2013, 16, 69–80.
- Bruce, T.J.A.; Matthes, M.C.; Napier, J.A.; Pickett, J.A. Stressful “memories” of plants: Evidence and possible mechanisms. Plant Sci. 2007, 173, 603–608.
- Pastor, V.; Luna, E.; Mauch-Mani, B.; Ton, J.; Flors, V. Primed plants do not forget. Environ. Exp. Bot. 2013, 94, 46–56.
- Shi, S.; Li, S.; Asim, M.; Mao, J.; Xu, D.; Ullah, Z.; Liu, G.; Wang, Q.; Liu, H. The Arabidopsis calcium-dependent protein kinases (CDPKs) and their roles in plant growth regulation and abiotic stress responses. Int. J. Mol. Sci. 2018, 19, 1900.
- Conrath, U. Molecular aspects of defence priming. Trends Plant Sci. 2011, 16, 524–531.
- Conrath, U.; Beckers, G.J.M.; Flors, V.; García-Agustín, P.; Jakab, G.; Mauch, F.; Newman, M.A.; Pieterse, C.M.J.; Poinssot, B.; Pozo, M.J.; et al. Priming: Getting ready for battle. Mol. Plant-Microbe Interact. 2006, 19, 1062–1071.
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013, 11, 745–760.
- Sahu, P.P.; Pandey, G.; Sharma, N.; Puranik, S.; Muthamilarasan, M.; Prasad, M. Epigenetic mechanisms of plant stress responses and adaptation. Plant Cell Rep. 2013, 32, 1151–1159.
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.M.; Bakker, P.A.H.M. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375.
- Mattoo, A.J.; Nonzom, S. Endophytic fungi: Understanding complex cross-talks. Symbiosis 2021, 1–28.
- Mendoza-Mendoza, A.; Zaid, R.; Lawry, R.; Hermosa, R.; Monte, E.; Horwitz, B.A.; Mukherjee, P.K. Molecular dialogues between Trichoderma and roots: Role of the fungal secretome. Fungal Biol. Rev. 2018, 32, 62–85.
- Ramírez-Valdespino, C.A.; Casas-Flores, S.; Olmedo-Monfil, V. Trichoderma as a model to study effector-like molecules. Front. Microbiol. 2019, 10, 1030.
- Alfiky, A.; Weisskopf, L. Deciphering Trichoderma–plant–pathogen interactions for better development of biocontrol applications. J. Fungi 2021, 7, 61.
- Navazio, L.; Baldan, B.; Moscatiello, R.; Zuppini, A.; Woo, S.L.; Mariani, P.; Lorito, M. Calcium-mediated perception and defense responses activated in plant cells by metabolite mixtures secreted by the biocontrol fungus Trichoderma atroviride. BMC Plant Biol. 2007, 7, 1–9.
- Omann, M.; Zeilinger, S. How a mycoparasite employs G-protein signaling: Using the example of Trichoderma. J. Signal Transduct. 2010, 2010, 1–8.
- Zhong, C.L.; Zhang, C.; Liu, J.Z. Heterotrimeric G protein signaling in plant immunity. J. Exp. Bot. 2019, 70, 1109–1118.
- Pandey, S. Heterotrimeric G-protein signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 2019, 70, 213–238.
- Tseng, Y.-H.; Rouina, H.; Groten, K.; Rajani, P.; Furch, A.C.U.; Reichelt, M.; Baldwin, I.T.; Nataraja, K.N.; Uma Shaanker, R.; Oelmüller, R. An endophytic Trichoderma strain promotes growth of its hosts and defends against pathogen attack. Front. Plant Sci. 2020, 11, 573670.
- Bailey, B.A.; Bae, H.; Strem, M.D.; Crozier, J.; Thomas, S.E.; Samuels, G.J.; Vinyard, B.T.; Holmes, K.A. Antibiosis, mycoparasitism, and colonization success for endophytic Trichoderma isolates with biological control potential in Theobroma cacao. Biol. Control 2008, 46, 24–35.
- Galletti, S.; Burzi, P.L.; Cerato, C.; Marinello, S.; Sala, E. Trichoderma as a potential biocontrol agent for Cercospora leaf spot of sugar beet. BioControl 2008, 53, 917–930.
- Guo, Y.; Ghirardo, A.; Weber, B.; Schnitzler, J.-P.; Benz, J.P.; Rosenkranz, M. Trichoderma species differ in their volatile profiles and in antagonism toward ectomycorrhiza Laccaria bicolor. Front. Microbiol. 2019, 10, 891.
- Oszako, T.; Voitka, D.; Stocki, M.; Stocka, N.; Nowakowska, J.A.; Linkiewicz, A.; Hsiang, T.; Belbahri, L.; Berezovska, D.; Malewski, T. Trichoderma asperellum efficiently protects Quercus robur leaves against Erysiphe alphitoides. Eur. J. Plant Pathol. 2021, 159, 295–308.
- Rocha, S.L.; Evans, H.C.; Jorge, V.L.; Cardoso, L.A.O.; Pereira, F.S.T.; Rocha, F.B.; Barreto, R.W.; Hart, A.G.; Elliot, S.L. Recognition of endophytic Trichoderma species by leaf-cutting ants and their potential in a Trojan-horse management strategy. R. Soc. Open Sci. 2017, 4, 160628.
- Kadota, Y.; Shirasu, K.; Zipfel, C. Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 2015, 56, 1472–1480.
- Morán-Diez, M.E.; Cardoza, R.E.; Gutiérrez, S.; Monte, E.; Hermosa, R. TvDim1 of Trichoderma virens is involved in redox-processes and confers resistance to oxidative stresses. Curr. Genet. 2010, 56, 63–73.
- Montero-Barrientos, M.; Hermosa, R.; Cardoza, R.E.; Gutiérrez, S.; Monte, E. Functional analysis of the Trichoderma harzianum nox1 gene, encoding an NADPH oxidase, relates production of reactive oxygen species to specific biocontrol activity against Pythium ultimum. Appl. Environ. Microbiol. 2011, 77, 3009–3016.
- Morán-Diez, E.; Hermosa, R.; Ambrosino, P.; Cardoza, R.E.; Gutiérrez, S.; Lorito, M.; Monte, E. The ThPG1 endopolygalacturonase is required for the Trichoderma harzianum-plant beneficial interaction. Mol. Plant-Microbe Interact. 2009, 22, 1021–1031.
- Alonso-Ramírez, A.; Poveda, J.; Martín, I.; Hermosa, R.; Monte, E.; Nicolás, C. Salicylic acid prevents Trichoderma harzianum from entering the vascular system of roots. Mol. Plant Pathol. 2014, 15, 823–831.
- Morán-Diez, E.; Rubio, B.; Domínguez, S.; Hermosa, R.; Monte, E.; Nicolás, C. Transcriptomic response of Arabidopsis thaliana after 24h incubation with the biocontrol fungus Trichoderma harzianum. J. Plant Physiol. 2012, 169, 614–620.
- Feys, B.J.; Moisan, L.J.; Newman, M.A.; Parker, J.E. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001, 20, 5400–5411.
- Salminen, A.; Kauppinen, A.; Kaarniranta, K. AMPK/Snf1 signaling regulates histone acetylation: Impact on gene expression and epigenetic functions. Cell. Signal. 2016, 28, 887–895.
- Shoresh, M.; Spivak, M.; Bernstein, N. Involvement of calcium-mediated effects on ROS metabolism in the regulation of growth improvement under salinity. Free Radic. Biol. Med. 2011, 51, 1221–1234.
- Chen, J.; Harman, G.E.; Comis, A.; Cheng, G.-W. Proteins related to the biocontrol of Pythium damping-off in maize with Trichoderma harzianum Rifai. J. Integr. Plant Biol. 2005, 47, 988–997.
- Yedidia, I.; Benhamou, N.; Chet, I. Induction of defense responses in cucumber plants (Cucumis sativus L.) by the biocontrol agent Trichoderma harzianum. Appl. Environ. Microbiol. 1999, 65, 1061–1070.
- De Palma, M.; D’Agostino, N.; Proietti, S.; Bertini, L.; Lorito, M.; Ruocco, M.; Caruso, C.; Chiusano, M.L.; Tucci, M. Suppression Subtractive Hybridization analysis provides new insights into the tomato (Solanum lycopersicum L.) response to the plant probiotic microorganism Trichoderma longibrachiatum MK1. J. Plant Physiol. 2016, 190, 79–94.
- Morán-Diez, M.E.; Tranque, E.; Bettiol, W.; Monte, E.; Hermosa, R. Differential response of tomato plants to the application of three Trichoderma species when evaluating the control of Pseudomonas syringae populations. Plants 2020, 9, 626.
- De Palma, M.; Salzano, M.; Villano, C.; Aversano, R.; Lorito, M.; Ruocco, M.; Docimo, T.; Piccinelli, A.L.; D’Agostino, N.; Tucci, M. Transcriptome reprogramming, epigenetic modifications and alternative splicing orchestrate the tomato root response to the beneficial fungus Trichoderma harzianum. Hortic. Res. 2019, 6, 1–15.
- Zehra, A.; Meena, M.; Dubey, M.K.; Aamir, M.; Upadhyay, R.S. Synergistic effects of plant defense elicitors and Trichoderma harzianum on enhanced induction of antioxidant defense system in tomato against Fusarium wilt disease. Bot. Stud. 2017, 58, 1–14.
- Aamir, M.; Kashyap, S.P.; Zehra, A.; Dubey, M.K.; Singh, V.K.; Ansari, W.A.; Upadhyay, R.S.; Singh, S. Trichoderma erinaceum bio-priming modulates the WRKYs defense programming in tomato against the Fusarium oxysporum f. sp. lycopersici (Fol) challenged condition. Front. Plant Sci. 2019, 10, 911.
- Herrera-Téllez, V.I.; Cruz-Olmedo, A.K.; Plasencia, J.; Gavilanes-Ruiz, M.; Arce-Cervantes, O.; Hernández-León, S.; Saucedo-García, M. The protective effect of Trichoderma asperellum on tomato plants against Fusarium oxysporum and Botrytis cinerea diseases involves inhibition of reactive oxygen species production. Int. J. Mol. Sci. 2019, 20, 2007.
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 2018, 9, 1836.
- Fernández, I.; Cosme, M.; Stringlis, I.A.; Yu, K.; Jonge, R.; van Wees, S.M.; Pozo, M.J.; Pieterse, C.M.J.; van der Heijden, M.G.A. Molecular dialogue between arbuscular mycorrhizal fungi and the nonhost plant Arabidopsis thaliana switches from initial detection to antagonism. N. Phytol. 2019, 223, 867–881.
- Xu, Y.; Zhang, J.; Shao, J.; Feng, H.; Zhang, R.; Shen, Q. Extracellular proteins of Trichoderma guizhouense elicit an immune response in maize (Zea mays) plants. Plant Soil 2020, 449, 133–149.
- Koornneef, A.; Pieterse, C.M.J. Cross talk in defense signaling. Plant Physiol. 2008, 146, 839–844.
- Carrero-Carrón, I.; Rubio, M.B.; Niño-Sánchez, J.; Navas-Cortés, J.A.; Jiménez-Díaz, R.M.; Monte, E.; Hermosa, R. Interactions between Trichoderma harzianum and defoliating Verticillium dahliae in resistant and susceptible wild olive clones. Plant Pathol. 2018, 67, 1758–1767.
- Beckers, G.J.M.; Jaskiewicz, M.; Liu, Y.; Underwood, W.R.; He, S.Y.; Zhang, S.; Conrath, U. Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 2009, 21, 944–953.
- Su, J.; Yang, L.; Zhu, Q.; Wu, H.; He, Y.; Liu, Y.; Xu, J.; Jiang, D.; Zhang, S. Active photosynthetic inhibition mediated by MPK3/MPK6 is critical to effector-triggered immunity. PLoS Biol. 2018, 16, e2004122.
- Pitzschke, A.; Schikora, A.; Hirt, H. MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 2009, 12, 421–426.
- Wiermer, M.; Feys, B.J.; Parker, J.E. Plant immunity: The EDS1 regulatory node. Curr. Opin. Plant Biol. 2005, 8, 383–389.
- Shoresh, M.; Gal-On, A.; Leibman, D.; Chet, I. Characterization of a mitogen-activated protein kinase gene from cucumber required for Trichoderma-conferred plant resistance. Plant Physiol. 2006, 142, 1169–1179.
- Contreras-Cornejo, H.A.; López-Bucio, J.S.; Méndez-Bravo, A.; Macías-Rodríguez, L.; Ramos-Vega, M.; Guevara-García, Á.A.; López-Bucio, J. Mitogen-activated protein kinase 6 and ethylene and auxin signaling pathways are involved in Arabidopsis root-system architecture alterations by Trichoderma atroviride. Mol. Plant-Microbe Interact. 2015, 28, 701–710.
- Liu, Y.; Zhang, S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 2004, 16, 3386–3399.
- Meng, X.; Xu, J.; He, Y.; Yang, K.Y.; Mordorski, B.; Liu, Y.; Zhang, S. Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 2013, 25, 1126–1142.
- Birkenbihl, R.P.; Diezel, C.; Somssich, I.E. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 2012, 159, 266–285.
- Mayo, S.; Cominelli, E.; Sparvoli, F.; González-López, O.; Rodríguez-González, A.; Gutiérrez, S.; Casquero, P.A. Development of a qPCR strategy to select bean genes involved in plant defense response and regulated by the Trichoderma velutinum–Rhizoctonia solani interaction. Front. Plant Sci. 2016, 7, 1109.
- Jung, H.W.; Tschaplinski, T.J.; Wang, L.; Glazebrook, J.; Greenberg, J.T. Priming in systemic plant immunity. Science 2009, 324, 89–91.
- Spoel, S.H.; Dong, X. How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 2012, 12, 89–100.
- Pitzschke, A.; Datta, S.; Persak, H. Salt stress in Arabidopsis: Lipid transfer protein AZI1 and its control by mitogen-activated protein kinase MPK3. Mol. Plant 2014, 7, 722–738.
- Segarra, G.; Van Der Ent, S.; Trillas, I.; Pieterse, C.M.J. MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant Biol. 2009, 11, 90–96.
- Mathys, J.; De Cremer, K.; Timmermans, P.; Van Kerckhove, S.; Lievens, B.; Vanhaecke, M.; Cammue, B.P.A.; De Coninck, B. Genome-wide characterization of ISR induced in Arabidopsis thaliana by Trichoderma hamatum T382 against Botrytis cinerea infection. Front. Plant Sci. 2012, 3, 108.
- Brotman, Y.; Landau, U.; Cuadros-Inostroza, Á.; Takayuki, T.; Fernie, A.R.; Chet, I.; Viterbo, A.; Willmitzer, L. Trichoderma-plant root colonization: Escaping early plant defense responses and activation of the antioxidant machinery for saline stress tolerance. PLoS Pathog. 2013, 9, e1003221.
- Sáenz-Mata, J.; Salazar-Badillo, F.B.; Jiménez-Bremont, J.F. Transcriptional regulation of Arabidopsis thaliana WRKY genes under interaction with beneficial fungus Trichoderma atroviride. Acta Physiol. Plant. 2014, 36, 1085–1093.
- Medeiros, H.A.; De Araújo Filho, J.V.; Freitas, L.G.; Castillo, P.; Rubio, M.B.; Hermosa, R.; Monte, E. Tomato progeny inherit resistance to the nematode Meloidogyne javanica linked to plant growth induced by the biocontrol fungus Trichoderma atroviride. Sci. Rep. 2017, 7, 1–13.
- Rubio, M.B.; Martínez de Alba, A.E.; Nicolás, C.; Monte, E.; Hermosa, R. Early root transcriptomic changes in wheat seedlings colonized by Trichoderma harzianum under different inorganic nitrogen supplies. Front. Microbiol. 2019, 10, 2444.
- Dou, K.; Wang, Z.; Zhang, R.; Wang, N.; Fan, H.; Diao, G. Cloning and characteristic analysis of a novel aspartic protease gene Asp55 from Trichoderma asperellum ACCC30536. Microbiol. Res. 2014, 169, 915–923.
- Yu, W.; Mijiti, G.; Huang, Y.; Fan, H.; Wang, Y.; Liu, Z. Functional analysis of eliciting plant response protein Epl1-Tas from Trichoderma asperellum ACCC30536. Sci. Rep. 2018, 8, 1–13.
- Banerjee, A.; Roychoudhury, A. WRKY proteins: Signaling and regulation of expression during abiotic stress responses. Sci. World J. 2015, 2015, 807560.
- Wang, D.; Amornsiripanitch, N.; Dong, X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2006, 2, e123.
- Zhang, Y.; Lee, C.-W.; Wehner, N.; Imdahl, F.; Svetlana, V.; Weiste, C.; Dröge-Laser, W.; Deeken, R. Regulation of oncogene expression in T-DNA-transformed plant cells. PLoS Pathog. 2015, 11, e1004620.
- Gao, X.; Cox, K.L.; He, P. Functions of calcium-dependent protein kinases in plant innate immunity. Plants 2014, 3, 160–176.
- Alvarez-Venegas, R.; Abdallat, A.A.; Guo, M.; Alfano, J.R.; Avramova, Z. Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics 2007, 2, 106–113.
- Jaskiewicz, M.; Conrath, U.; Peterhälnsel, C. Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 2011, 12, 50–55.
- Phukan, U.J.; Jeena, G.S.; Shukla, R.K. WRKY transcription factors: Molecular regulation and stress responses in plants. Front. Plant Sci. 2016, 7, 760.
- Barco, B.; Clay, N.K. Hierarchical and dynamic regulation of defense-responsive specialized metabolism by WRKY and MYB transcription factors. Front. Plant Sci. 2020, 10, 1775.
- Shim, J.S.; Choi, Y.D. Direct regulation of WRKY70 by AtMYB44 in plant defense responses. Plant Signal. Behav. 2013, 8, e24509.
- Shin, R.; Burch, A.Y.; Huppert, K.A.; Tiwari, S.B.; Murphy, A.S.; Guilfoyle, T.J.; Schachtman, D.P. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 2007, 19, 2440–2453.
- Van der Ent, S.; Verhagen, B.W.M.; Van Doorn, R.; Bakker, D.; Verlaan, M.G.; Pel, M.J.C.; Joosten, R.G.; Proveniers, M.C.G.; Van Loon, L.C.; Ton, J.; et al. MYB72 is required in early signaling steps of rhizobacteria-induced systemic resistance in Arabidopsis. Plant Physiol. 2008, 146, 1293–1304.
- Kranz, H.D.; Denekamp, M.; Greco, R.; Jin, H.; Leyva, A.; Meissner, R.C.; Petroni, K.; Urzainqui, A.; Bevan, M.; Martin, C.; et al. Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 1998, 16, 263–276.
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703.
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78.
- Zhai, Q.; Li, C. The plant Mediator complex and its role in jasmonate signaling. J. Exp. Bot. 2019, 70, 3415–3424.
- Çevik, V.; Kidd, B.N.; Zhang, P.; Hill, C.; Kiddle, S.; Denby, K.J.; Holub, E.B.; Cahill, D.M.; Manners, J.M.; Schenk, P.M.; et al. MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 2012, 160, 541–555.
- Caarls, L.; Van Der Does, D.; Hickman, R.; Jansen, W.; Van Verk, M.C.; Proietti, S.; Lorenzo, O.; Solano, R.; Pieterse, C.M.J.; Van Wees, S.C.M. Assessing the role of ETHYLENE RESPONSE FACTOR transcriptional repressors in salicylic acid-mediated suppression of jasmonic acid-responsive genes. Plant Cell Physiol. 2017, 58, 266–278.
- Liu, Y.; Du, M.; Deng, L.; Shen, J.; Fang, M.; Chen, Q.; Lu, Y.; Wang, Q.; Li, C.; Zhai, Q. MYC2 regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop. Plant Cell 2019, 31, 106–127.
- Wang, H.; Li, S.; Li, Y.; Xu, Y.; Wang, Y.; Zhang, R.; Sun, W.; Chen, Q.; Wang, X.J.; Li, C.; et al. MED25 connects enhancer–promoter looping and MYC2-dependent activation of jasmonate signalling. Nat. Plants 2019, 5, 616–625.
- Chen, R.; Jiang, H.; Li, L.; Zhai, Q.; Qi, L.; Zhou, W.; Liu, X.; Li, H.; Zheng, W.; Sun, J.; et al. The Arabidopsis Mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 2012, 24, 2898–2916.
- Raya-González, J.; Ortiz-Castro, R.; Ruiz-Herrera, L.F.; Kazan, K.; López-Bucio, J. Phytochrome and flowering time1/mediator25 regulates lateral root formation via auxin signaling in Arabidopsis. Plant Physiol. 2014, 165, 880–894.
- Peñuelas, M.; Monte, I.; Schweizer, F.; Vallat, A.; Reymond, P.; García-Casado, G.; Franco-Zorrilla, J.M.; Solano, R. Jasmonate-related MYC transcription factors are functionally conserved in Marchantia polymorpha. Plant Cell 2019, 31, 2491–2509.




