
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | ZEYNETTIN AKKUS | + 1184 word(s) | 1184 | 2021-04-28 09:00:37 | | | |
| 2 | Lindsay Dong | Meta information modification | 1184 | 2021-04-28 09:37:00 | | |
Video Upload Options
Echocardiography (Echo), a widely available, noninvasive, and portable bedside imaging tool, is the most frequently used imaging modality in assessing cardiac anatomy and function in clinical practice. Artificial-intelligence-empowered echo (AI-Echo) can potentially reduce inter-interpreter variability and indeterminate assessment and improve the detection of unique conditions as well as the management of various cardiac disorders.
1. Introduction
Echocardiography (Echo), also known as cardiac ultrasound (CUS), is currently the most widely used noninvasive imaging modality for assessing patients with various cardiovascular disorders. It plays a vital role in evaluation of patients with symptoms of heart disease by identifying structural as well as functional abnormalities and assessing intracardiac hemodynamics. However, accurate echo measurements can be hampered by variability between interpreters, patients, and operators and image quality. Therefore, there is a clinical need for standardized methods of echo measurements and interpretation to reduce these variabilities.
1.1. Transthoracic Echocardiogram
Transthoracic echocardiogram transmits and receives sound waves with frequencies higher than human hearing using an ultrasound transducer. It generates ultrasound waves and transmits to the tissue and listens to receive the reflected sound wave (echo). CUS has several advantages compared to cardiac magnetic resonance, cardiac computed tomography, and cardiac positron emission tomography imaging modalities. CUS does not use ionizing radiation, is less expensive, portable for point-of-care (POCUS) applications, and provides actual real-time imaging.
1.2. Artificial Intelligence
Artificial intelligence (AI) is considered to be a computer-based system that can observe an environment and takes actions to maximize the success of achieving its goals. Some examples include a system that has the ability of sensing, reasoning, engaging, and learning, are computer vision for understanding digital images, natural language processing for interaction between human and computer languages, voice recognition for detection and translation of spoken languages, robotics and motion, planning and organization, and knowledge capture. ML is a subsection of AI that covers the ability of a system to learn about data using supervised or unsupervised statistical and ML methods such as regression, support vector machines, decision trees, and neural networks. Deep learning (DL), which is a subclass of ML, learns a sequential chain of pivotal features from input data that maximizes the success of the learning process with its self-learning ability. This is different from statistical ML algorithms that require handcrafted feature selection [1] (Figure 1).
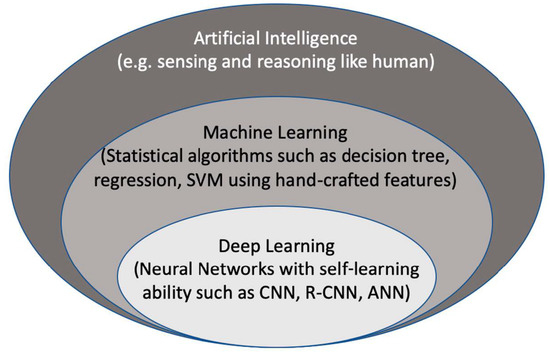
Figure 2. The context of artificial intelligence, machine learning, and deep learning. SVM: Support Vector Machine. CNN: convolutional neural networks, R-CNN: recurrent CNN, ANN: artificial neural networks.
The representation power of DL models is massive and can create representation for any given variation of a signal. Recent accomplishments of DL, especially in image classification and segmentation applications, made it very popular in the data science community. Traditional ML methods use hand-crafted features extracted from data and process them in decomposable pipelines. This makes them more comprehensible as each component is explainable. On the other hand, they tend to be less generalizable and robust to variations in data.
2. Artificial Intelligence (AI)-Empowered Echocardiography Interpretation
Automated image interpretation that mimics human vision with traditional machine learning has existed for a long time. Recent advances in parallel processing with GPUs and deep-learning algorithms, which extract patterns in images with their self-learning ability, have changed the entire automated image interpretation practice with respect to computation speed, generalizability, and transferability of these algorithms. AI-empowered echocardiography has been advancing and moving closer to be used in routine clinical workflow in cardiology due to the increased demand for standardizing acquisition and interpretation of cardiac US images. Even though DL-based methods for echocardiography provide promising results in diagnosis and quantification of diseases, AI-Echo still needs to be validated with larger study populations including multi-center and multi-vendor datasets. High intra-/inter-variability in echocardiography makes standardization of image acquisition and interpretation challenging. However, AI-Echo will provide solutions to mitigate operator-dependent variability and interpretability. AI applications in cardiac US are more challenging than those in cardiac CT and MR imaging modalities due to patient-dependent factors (e.g., obesity, limited acoustic window, artifacts, and signal drops) and natural US speckle noise pattern. These factors that affect US image quality will remain as challenges with cardiac ultrasound.
Applications of DL in echocardiography are rapidly advancing as evidenced by the growing number of studies recently. DL models have enormous representation power and are hungry for large amounts of data in order to obtain generalization ability and stability. Creating databases with large datasets that are curated and have good quality data and labels is the most challenging and time-consuming part of the whole AI model development process. Although it has been shown that AI-echo applications have superb performance compared to classical ML methods, most of the models were trained and evaluated on small datasets. It is important to train AI models on large multi-vendor and multi-center datasets to obtain generalization and validate on large multi-vendor datasets to increase reliability of a proposed model. An alternative way to overcome the limitation of having small training datasets would be augmenting the dataset with realistic transformations (e.g., scaling, horizontal flipping, translations, adding noise, tissue deformation, and adjusting image contrast) that could help improve generalizability of AI models. On the other hand, realistic transformations need to be used to genuinely simulate variations in cardiac ultrasound images, and transformations-applied images should not create artifacts. Alternatively, generative adversarial networks, which include a generator and a discriminator model, are trained until the model generates images that are not separable by the discriminator. This could be used to generate realistic cardiac ultrasound B-mode images of the heart. Introducing such transformations during the training process will make AI models more robust to small perturbations in input data space.
It is important to design AI models that are transparent for the prediction of any disease from medical images. The AI models developed for diagnosis of a disease must elucidate the reasons and motivations behind their predictions in order to build trust in them. Comprehension of the inner mechanism of an AI model necessitates interpreting the activity of feature maps in each layer [2][3][4]. However, the extracted features are a combination of sequential layers and become complicated and conceptual with more layers. Therefore, the interpretation of these features become difficult compared to handcrafted imaging features in traditional ML methods. Traditional ML methods are designed for separable components that are more understandable, since each component of ML methods has an explanation but usually is not very accurate or robust. With DL-based AI models, the interpretability is given up for the robustness and complex imaging features with greater generalizability. Recently, a number of methods have been introduced about what DL models see and how to make their predictions. Several CNN architectures [5][6][7][8][9] employed techniques such as deconvolutional networks [10], gradient back-propagation [11], class activation maps (CAM) [4], gradient-weighted CAM [12], and saliency maps [13][14] to make CNN understandable. With these techniques, gradients of a model have been projected back to the input image space, which shows what parts in the input image contribute the most to the prediction outcome that maximizes the classification accuracy. Although making AI models understandable has been an active research topic in the DL community, there is still much further research needed in the area.
References
- Akkus, Z.; Galimzianova, A.; Hoogi, A.; Rubin, D.L.; Erickson, B.J. Deep Learning for Brain MRI Segmentation: State of The Art and Future Directions. J. Digit. Imaging 2017, 30, 449–459.
- Zeiler, M.D.; Fergus, R. Visualizing and Understanding Convolutional Networks. In Proceedings of the Lecture Notes in Computer Science, Zurich, Switzerland, 6–12 September 2014; pp. 818–833.
- Zeiler, M.D.; Taylor, G.W.; Fergus, R. Adaptive deconvolutional networks for mid and high level feature learning. In Proceedings of the 2011 International Conference on Computer Vision, Washington, DC, USA, 6–13 November 2011.
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning Deep Features for Discriminative Localization. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016.
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556.
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015; Lecture Notes in Computer Science; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Munich, Germany, 2015; Volume 9351, pp. 234–241. ISBN 9783319245737.
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385.
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826.
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495.
- Zeiler, M.D.; Krishnan, D.; Taylor, G.W.; Fergus, R. Deconvolutional Networks; IEEE Computer Society: San Francisco, CA, USA, 2010; pp. 2528–2535. ISBN 9781424469840.
- Springenberg, J.T.; Dosovitskiy, A.; Brox, T.; Riedmiller, M. Striving for Simplicity: The All Convolutional Net. arXiv 2014, arXiv:1412.6806.
- Chattopadhay, A.; Sarkar, A.; Howlader, P.; Balasubramanian, V.N. Grad-CAM++: Generalized Gradient-Based Visual Explanations for Deep Convolutional Networks. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; pp. 839–847.
- Li, G.; Yu, Y. Visual Saliency Detection Based on Multiscale Deep CNN Features. IEEE Trans. Image Process. 2016, 25, 5012–5024.
- Philbrick, K.A.; Yoshida, K.; Inoue, D.; Akkus, Z.; Kline, T.L.; Weston, A.D.; Korfiatis, P.; Takahashi, N.; Erickson, B.J. What Does Deep Learning See? Insights from a Classifier Trained to Predict Contrast Enhancement Phase from CT Images. AJR Am. J. Roentgenol. 2018, 211, 1184–1193.





