
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ahlem Teniou | + 4573 word(s) | 4573 | 2021-04-14 08:33:32 | | | |
| 2 | Conner Chen | -244 word(s) | 4329 | 2021-04-21 04:45:12 | | |
Video Upload Options
To date, six human coronaviruses have been identified: α-coronaviruses (HCoVs-NL63, HCoVs-229E), β-coronaviruses (HCoVs-OC43, HCoVs-HKU1), severe acute respiratory syndrome-CoV (SARS-CoV), and Middle East respiratory syndrome-CoV (MERS-CoV). After the SARS-CoV-1 epidemic, the world is living a new threat to human health since December 2019—the SARS-CoV-2 or the COVID-19 pandemic. The emergence of the novel coronavirus is associated with an atypical pneumonia that has led to 90,176,569 infections and 1,936,617 deaths worldwide, as of 10 January 2021. Structurally, SARS-CoV-2 is an enveloped RNA(Ribonucleic acid) virus comprising a spike protein (S), a hemagglutinin-esterase dimer (HE), a membrane glycoprotein (M), an envelope protein (E), and a nucleocapsid protein (N). It has been demonstrated that the mechanism of the viral infection requires angiotensin-converting enzyme 2 (ACE2) binding to the protein S with high affinity. Highly expressed in the endothelial cells of the cardiovascular system and kidneys, this human receptor is used by the virus as an entry to invade target cells.
Currently, immunoassays are the most popular diagnostic tools available in the market and used in medical structures. Basically, these methods use antibodies as bioreceptors targeting capsid proteins or whole viruses. In serological testing, capsid proteins are used as viral antigens to bind the immunoglobulins generated by the patient against the pathogen. Antibodies are usually obtained from animal immunization with N, S, or E protein or from the blood samples of patients who are infected [14]. In addition to the commercialized ELISA kits and rapid tests, several research reports have described novel immunoassays and immunosensors for coronavirus detection. We discuss in this part the principle of these methods as well as the most important results.
1. Electrochemical Immunosensors
Electrochemical immunosensing is based on the conversion of the antibody–antigen interaction into electrochemical signals. In contrast to traditional immunoassays, innovative transducers allow a highly sensitive quantification of the targeted antigen/antibody in different ways [1]. It should be noted that sensitivity plays a crucial role in viruses detection; lowering the detection limit allows the early diagnosis of the infection before the first symptoms [2]. In addition to their sensitivity and robustness, electrochemical devices can be miniaturized, providing an immediate response without referring to specialized labs and qualified staff [3]. For instance, Fabiani et al. employed carbon-black-based screen-printed electrodes as transducers and magnetic beads as an immobilization support for the immunological chain comprising anti-IgG (Immunoglobulin G) bound to the anti-S and anti-N antibodies. The principle is based on the specific recognition of the anti-S and/or anti-N antibodies with the virus present in the sample. After the antibody–antigen interaction, polyclonal anti-S and anti-N antibodies are added, followed by an alkaline-phosphatase-labeled secondary antibody. Finally, the conversion of the enzymatic substrate to the electro-active by-product 1-naphtol was monitored by differentialpulse voltammetry (DPV) using a portable potensiostat connected to a computer. The immunosensor analytical performances were confirmed in standard solutions of S and N proteins and untreated saliva. Moreover, experiments using cultured virus at biosafety level 3 and in saliva clinical samples were carried out by comparing the data using the nasopharyngeal swab specimens tested with RT-PCR. Despite its high sensitivity for SARS-CoV-2 and selectivity toward H1N1 influenza virus, the reported immunosensor requires several steps of construction and antibodies, thusincreasing the complexity and cost of the method [4]. Later, a simple and low-cost electrochemical immunosensing strategy was reported for the simultaneous measurement of the nucleocapsid protein, IgG and IgM (Immunoglobulin M) specific to the S1 protein, as well as C-reactive protein (CRP) levels. These biomarkers provide information about the infection, immune response, and disease severity. The telemedicine Rapidplex platform is composed of four graphene electrodes fabricated by laser engraving [5]. Electrodes are designed based on the sandwich/double sandwich for Igsand indirect immunosensing for N and CRP (Figure 1). The effectiveness of each electrode was monitored amperometrically in blood and saliva samples. Excellent results were obtained in terms of sensitivity, selectivity toward SARS-CoV-1 and MERS-CoV, as well as reproducibility and stability [6]. In brief, this innovative platform should meet the high requirement for modern diagnosis tools allowing accurate, simple and rapid monitoring of SARS-CoV2 infection. In a recent study, Zourob’s group developed cotton tipped electrochemical immunosensor for the nucleocapsid protein detection. The novelty of the described strategy is in combing the sample collection and detection tools in a single platform by coating screen-printed electrodes with absorbing cotton padding. The detection principle was based on the competition between the immobilized antigen on screen printed electrodes and that present in nasal samples, to bind N protein antibody in solution. This platform would avoid sample pre-treatment and transfer which are indispensable in the other diagnosis approaches [7].
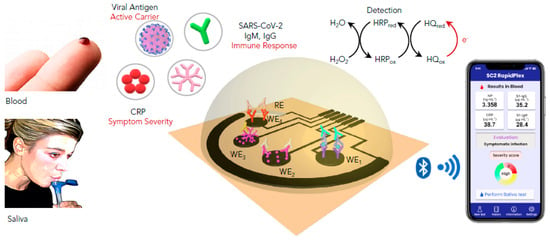
Figure 1. Schematic representation of the graphene-based electrochemical telemedicine platform for multiplexed detection of CRP, (SARS-CoV2) antigens, IgG (Immunoglobulin G) and IgM (Immunoglobulin M). This RapidPlex is composed of four graphene working electrodes (WEs); Ag/AgCl reference electrode (RE), and a graphene counter electrode (CE). All of them patterned on a polyimide (PI) substrate via CO2 laser engraving, and a fast, high-throughput. Data can be wirelessly transmitted to a mobile user interface.Reprinted with permission from [6].
2. Field-Effect Transistor-Based Immunosensors
Field-effect transistor(FET)-based biosensing is characterized by many advantages, including sensitivity, fast response, and low-noise detection [8]. Moreover, FET-based biosensing does not require labeling of bioreceptors and targets, making the strategy simpler and cheaper. Therefore, this strategy represents great potential in point-of-care testing and clinical diagnosis of viral infections. Aiming to detect the new coronavirus with high sensitivity, Zhang et al. developed a graphene-FET immunosensor for electrical probing of the spike protein S1 [9]. Graphene is widely used in biosensing platforms because of its remarkable conductivity, carrier mobility, and large surface area [10]. In this work, the subunit S1 was targeted because it is less conserved than N and S2 proteins in SARS-CoV [11]. Furthermore, the S1 protein contains a receptor-binding domain (RBD), which interacts with the ACE2 receptor [12]. In brief, the reported immunosensor was constructed by functionalizing the graphene surface with the S1-specific antibody or ACE2. The formation of the complex antibody/ACE-S1 subunit directly alters the conductance/resistance via field effect, resulting in a measurable electrical signal. The developed platform could provide an alternative platform for early screening and diagnosis. It would be also useful in rational design of neutralizing antibodies and docking methods as well as developing vaccines, prophylactics and therapeutics to combat COVID-19 [9]. Despite the high sensitivity of the reported immunosensor, no results of clinical sample application were reported. In another study, a highly sensitive FET-based immunosensor was constructed and successfully applied for spike protein detection in cultured virus and nasopharyngeal swab specimens from COVID-19 patients [13].
3. Optical Immunosensors
Besides electrochemical methods, optical biosensors have been widely used in clinical diagnosis and drug discovery due to their analytical performance in detecting biological systems [14]. Different approaches have been reported for optical immunosensing of the novel coronavirus, including surface plasmon resonance (SPR), chemiluminescence, and colorimetry.
Surface plasmon resonance-based biosensors are based on the monitoring of refractive index variations after the formation of the target–receptor complex on the sensing surface. Plasmonic biosensing offers many advantages, including label-free and real-time monitoring [15]. Yong’s group described a sandwich plasmonic immunosensor to detect a spike protein in serum samples. The principle of the assay is based on functionalizing a gold nanosheet with the protein S specific antibody by conjugating gold nanorods to the same antibody for sensitivity enhancement. Then, samples containing the antigen were allowed to flow as an analyte. This work was assisted by a simulation technique to study the effect of ARs and arrangements of the gold nanorods [16].
Besides label-free sensors, optical sensors, based on chemiluminescent and colorimetric labeling, have been reported for COVID-19 diagnosis. First, Cai et al. developed a chemiluminescent enzymatic serological immunoassay based on a peptide from the S protein. The latter was screened from peptide antigens synthesized from the genomic sequence from GenBank (NC_045512.1). Streptavidin-functionalized magnetic beads were used as an immobilization support for the biotinylated peptide. The binding between IgM and IgG present in serum samples and the magnetic beads was carried out in solution. After binding and washing, the conjugate was reacted with the substrate, generating a luminescent signal, which was measured by a luminometer. In this work, no clinical application was carried out [17]. Another research group reported, subsequently, the development and clinical application of a colorimetric qualitative lateral flow immunoassay for simultaneous detection of IgM and IgG in blood. The rapid test is composed of anti-IgG and anti-IgM antibodies stripped on two separate test lines. In parallel, a conjugate of viral antigen-AuNPs is sprayed on conjugation pads to generate a colorimetric signal. The presence of IgG and/or IgM antibodies is indicated by a red/purple line. The rapid test was applied on 397 PCR-confirmed patients and 128 negative patients at eight different clinical sites, showing an overall testing sensitivity of 88.66% and a specificity of 90.63% [18].
Despite the importance of IgGs in detecting previous COVID-19 infection and the association of IgM to recent contamination, the role of secretory immunoglobulins IgA should not be neglected. IgAs(Immunoglobulin A)play a key role in the immune exclusion process limiting the access of microorganisms to the mucosal barriers. It has been demonstrated that elevated total IgA and IgA-aPL(IgA-antiphospholipid) are significantly associated with severe COVID-19 infection [19][20]. Roda et al. developed a dual optical immunosensing platform for IgA detection in saliva by using the nucleocapsid protein as a receptor. The anti-human IgA was labeled with AuNPs for colorimetric detection, while HRP(HorsradishPeroxidase)was used to generate chemiluminescence after reaction with the luminol substrate. IgA present in serum/saliva is recognized by the immobilized N protein, and the secondary antibody captures the formed complex, thus generating a colorimetric/chemiluminescent signal. For colorimetry, a smartphone camera was used to capture the colored strip, while the light emitted by the chemiluminescence reaction (luminol-HRP) was measured by a cooled CCD (Charge Coupled Device) in contact imaging mode, with the data reported in relative light units. In contrast to the previously discussed LFIA (Lateral Flow Immunoassay), the one described in this report provides a qualitative and quantitative analysis. The use of this rapid test would assist studies on the role of IgA in the SARS-CoV-2 [21]. In addition to immunoglobulins, rapid tests based on LFIA have been also reported for capsid protein detection. For instance, Diao et al. developed a fluorescent immunoassay to detect a nucleocapsid protein in nasopharyngeal swab samples and urine within 10 min [22].
The different immunosensing strategies described above are summarized in Table 1 and Table 2.
Table 1. Electrochemical biosensors for SARS-CoV-2 detection.
| Detectionmethod | Target Genes | LOD | Time | Portability | Ref |
|---|---|---|---|---|---|
| Electricalconductivitychanges | S protein | 2.25 × 10−6 μg/mL | 10–30 s | Portable | [23] |
| FET | S protein | 10−9 μg/mL | Real time | Nonportable | [13] |
| Gr-FET | S1 protein (RBD) | <1.538 × 10−4 μg/mL | 2 min | Nonportable | [9] |
| Differential pulse voltammetry (DPV) | S protein / N protein | Sprotein: 19 × 10−3 μg/mL Nprotein: 8 × 10−3 μg/mL |
30 min | Portable | [4] |
| Impedancespectroscopy (EIS) | SARS-CoV-2 antibodies | Not available | <5 min | Portable | [24] |
| DPV | SARS-CoV-2 S-glycoproteins | 1.68 × 10−22 μg/mL | 1 min | Nonportable | [25] |
| DPV+ OCP-EIS | S1-IgG, S1-IgM, NP, CRP | Not available | 1 min | Portable | [6] |
| Square-wavevoltammetry | SARS-CoV-2 antigen | 0.8 pg/mL | Unknown | Nonportable | [7] |
FET: Field Effect Transistor, RBD (Receptor Binding Domain), CRP (C-Reactive Protein).
Table 2. Optical biosensors for SARS-CoV-2 detection.
| Detectionmethod | Target Genes | LOD | Time | Portability | Ref |
|---|---|---|---|---|---|
| Surface plasmonresonance (SPR) | Nucleocapsidof anti-SARS-CoV-2 antibodies | Not available | 15 min | Portable | [26] |
| LFIA | IgM and IgGantibodies | Not available | 15 min | Portable | [18] |
| LFIA | SARS-CoV-2 NP antigen | 250 pfu/µL | 20 min | Portable | [27] |
| LFIA | S1 protein | 1.86 × 105 copies/mL | 20 min | Portable | [28] |
| Colloidal gold-nanoparticle-basedLFIA | IgM antibodies | Not available | 15 min | Portable | [29] |
| SERS-LFIA | Anti-SARS-CoV-2 IgM/IgG | Not available | Real time | Portable | [30] |
| ELISA+GICA | IgM and IgG | Not available | GICA: 10min | Nonportable | [31] |
| Grating-coupledfluorescentplasmonic | Antibodiesagainst RBD; spike S1 fragment; spike S1 S2 extracellulardomain, andNprotein | Not available | <30 min | Nonportable | [32] |
| Immunochromatographicstrip | IgM orIgGantibody | Not available | 15 min | Portable | [33] |
| Fluorescenceimmunochromatographic | N protein | Not available | 10 min | Portable | [22] |
| Nanoplasmonicresonancesensor | S protein | 0.37 copies/µL | <15 min | Portable | [34] |
| Microfluidicimmunoassay-basedfluorescence | IgG/IgM/Antigen of SARS-CoV-2 | Not available | <15 min | Portable | [35] |
| Peptide-basedluminescentimmunoassay | IgG and IgM | Not available | Unknown | Nonportable | [17] |
| rN-based and rS-basedELISAs | IgM and IgGantibodies | Not available | Unknown | Nonportable | [36] |
| RT-LAMP-NBS | F1ab and N protein genes | 12 copies/µL | ≈1 h | Portable | [37] |
| AuNR-based SPR | S protein | Not available | Fewminutes | Nonportable | [16] |
| LFIA | Anti-SARS-CoV-2 IgA | Not available | 15 min | Portable | [21] |
PFU (plaque-forming unit): a measure used invirologyto describe the number of virus particles capable of forming plaques per unit volume, LFIA (Lateral Flow immunoassay), ELISA (Enzyme Linked Immunosorbent Assay), IgG (Immunoglobulin G), IgM (Immunoglobulin M), NP (Nucleocapsid protein).
4. Nucleic Acid-Based Techniques
As coronaviruses are positive single-stranded RNA viruses, their detection is usually based on nucleic acid techniques, mainly real-time PCR. However, this technology requires expensive reagents and equipment, in addition to qualified manipulators. Therefore, researchers are focusing on new techniques based on clustered regularly interspaced short palindromic repeats (CRISPR), aptamers, and nucleic acidhybridization to reduce the dependence on RT-PCR.
4.1. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Based Biosensing
After the identification of CRISPR-Cas systems targeting RNA, numerous studies have shown the potential use of this tool in the detection of different viruses such as dengue and Zika viruses [38]. Cas13 effector proteins are able to target RNA, with two RNase activities: gRNA maturation and both cis- and trans-RNA target cleavage [39]. They hold, thus, great promise for SARS-CoV-2 diagnosis and treatment. CRISPR-based platforms provide sensitive, rapid, and low-cost virus detection. Moreover, CRISPR-based diagnosis is usually based on isothermal amplification techniques that can be performed without thermocycler (recombinase polymerase amplification (RPA) and loop-mediated isothermal amplification (LAMP)). Several platforms have been applied for COVID-19 detection: specific high-sensitivity enzymatic reporter unlocking (SHERLOCK), HOLMES (one-hour low-cost multipurpose highly efficient system), DNA (Desoxyribonucleic acid), endonuclease-targeted CRISPR trans reporter (DETECTR), CAS-EXPAR(EXPonantial Amplification Reaction), NASBACC (nucleic acid sequences-based amplification CRISPR Cleavage), STOPCovid, ctPCR (cycle threshold PCR), and AIOD-CRISPR(All in one dual crispr). These platforms have been well discussed in a critical review by Vatankhah et al. [40].
The first designed and most used platform, called SHERLOCK showed a successful application for identifying cell-free tumor DNA mutations, genotyping human DNA, detectingdengue and Zika viruses, and distinguishing pathogenic bacteria [40]. Several studies have also demonstrated the potential application of this platform for COVID-19 detection [41]. SHERLOCK is simply based on Cas13a activation by the binding and cleavage of the target RNA. As a result, the nontarget molecules will be degraded including the quenched fluorescent reporter molecules. The released signal allows thus detection of the specific RNA after degradation of the non-targeted one [38]. Based on this principle, Zhang et al reported the first CRISPR-Cas13-based technique for COVID-19 detection by targeting S and ORF1ab protein genes. The presence of coronavirus in the sample activates the cas13 cleavage system by generating a measurable fluorescent signal. The developed technique allows the quantification of the coronavirus target genes in the range (20–200 aM) corresponding to (10–100 copies/µL). The whole test can be performed using a dipstick in less than one hour [42]. CRISPR-Cas13 technology can be thus considered as an excellent alternative to qRT-PCR for coronavirus diagnosis. In addition, the FDA (Food and Drug Administration) authorized the use of the Sherlock CRISPR SARS-CoV-2 Kit proposed by Sherlock BioSciencesCompany in May, 2020 [43] Subsequently, the clinical validation of this test has been reported by Patchsung et al in 154 nasopharyngeal and throat swab samples collected at Siriraj Hospital, Thailand [44].
By combining SHERLOCK with the HUDSON protocol; a process for lysis of viral particles and inactivation of DNases and RNases in body fluids with heat and chemical treatment, no prior RNA extraction is needed [40]. In a recent study, HUDSON was improved to allow virus inactivation within 10 minthusproviding a simple, rapid and single step Cas-13 detection tool of COVID-19. The reported test can be used outside hospitals and clinical laboratories [45].
Apart from Cas-13, Cas-12 effectors have been also employed for coronavirus diagnosis. For that, CRISPR-Cas12 was combined to DETCTR platform with the DNA endonuclease-targeted CRISPR trans reporter (DETCTR) platform. Extracted RNA is first retro-transcripted to DNA, followed by isothermal amplification using the RT-RPA enzyme. The Cas12a-gRNA complex is then activated through identification of a specific sequence in the amplified DNA, leading to the cleavage of reporters as it was described for the SHERLOCK platform. Based on this principle, different fluorescent lateral flow assays have been developed and tested in clinical samples, showing excellent results [46].
CRISPR-Cas9 effectors have been also exploited to construct a triple line lateral flow assay for the simultaneous dual gene (E and Orf1ab) detection of SARS-Cov2. The assay is based on the recognition between the Cas9/sgRNA and DNA-AuNPs probes. After binding, the formed complex accumulates on the test line and generates a visible colorimetric signal. The originality of this strip is the simultaneous detection of more than one gene which is not available by using Cas13 and 12 requiring nonspecific cleavage [47].
Finally, these studies confirm that CRISPR technology holds a great potential for the diagnosis of the novel coronavirus. It constitutes a promising alternative for the RT-qPCR technique, mainly due to its low cost, simple fabrication and sensitivity.
4.2. Aptamer-Based Biosensing
Aptamers are a class of bio-inspired receptors composed of single stranded DNA or RNA. They are selected by a combinatorial process called selection of ligands by exponantial enrichment (SELEX)for their affinity and specificity to a target [48]. They are able to bind their targets with high affinity and specificity comparable to antibodies. In addition, they are characterized by many advantages compared to antibodies, including cheap and simple chemical synthesis as well as stability in temperature and pH variations [49]. Moreover, aptamers can be easily functionalized with various chemical groups and labels allowing their use in different analytical applications [50].
In April 2020, Song et al selected the first aptamer specific for receptor binding domain of the SARS-CoV-2 spike glycoprotein. SELEX was carried out by incubating a ssDNA (single stranded DNA) library comprising 40 random nucleotides with RBD protein immobilized on A-beads. After a total of twelve rounds of selection and amplification, two sequences have been chosen. They exhibited a high affinity for the novel coronavirus with respective Kd values of 5.8 and 19.9 nM. In addition to the experimental competition, molecular dynamic simulations suggested that the selected aptamers bind to identical amino acids of RBD, thus providing a promising issue for coronavirus diagnosis, prevention and treatment [51]. The feasibility of the selected aptamer for spike protein detection has been subsequently, investigated by another research group. For that, the biosensor platform was constructed by immobilizing the amino-modified S-specific aptamer on a silicon thin film transistor. A wide range of potentials was applied to obtain a broader concentration dependency and study the binding mechanism between the aptamer and the spike protein. As compared to the FET-based immunosensor discussed above [52] the FET-based aptasensor showed a similar response concerning for S protein detection; the ratio increased by approximately 10% at the maximum spike concentration compared to the 0% response [52]. Therefore, this study confirms the potential application of the aptamer targeting the S protein in COVID-19 accurate diagnosis.
Apart from the spike protein, Nucleocapsid protein (NP) has been also targeted by SELEX technology. Zhang et al selected four aptamers specific for N protein with high affinity lower than 5 nM.For that, a total of five rounds of selection were performed starting from a library of 76 nucleotides comprising a randomized sequence of 36 nucleotides. After being selected, aptamers were employed in a sandwich-type assay based on four aptamer pairs, which form a ternary complex with NP. Combined with antibodies, this principle was first applied in an enzyme linked immunosorbent assay showing a high sensitivity. Then, an immunochromatographic strip was developed using gold nanoparticles to allow the visual detection of NP in urine and serum [53]. The selected aptamers hold a great promise for aptamer-based diagnosis and treatment of SARS-CoV-2. However, the developed tests require pairs of aptamers and antibodies thus increasing the device cost and complexity.
Besides to aptamers specifically selected for the novel coronavirus, the potential use of SARS-CoV-1 specific aptamers has been also investigated. Based on the fact that the sequence of SARS-CoV-2 N protein exhibits a homology of 91% with that of SARS-CoV, Chen et al studied the binding capacities of the SARS-CoV aptamer to detect the new coronavirus. Enzyme-linked aptamer binding assay (ELAA) demonstrated that the three tested aptamers exhibit a binding affinity for the SARS-CoV-2 N protein opening the way for their application in COVID-19 diagnosis and treatment. Aiming to validate that, the binding efficiency of aptamer 2 was compared to that of a commercial antibody using Western blot showing similar results. These aptamers could thus be a good alternative for antibodies and immunoassays, currently used for coronavirus detection. Moreover, the use of these aptamers should provide a low cost and rapid solution by avoiding the selection of new candidates specific to SARS-CoV-2 [54].
Recently, an interesting study was published by exploring the binding affinity of a nucleocapsid protein specific aptamer and proximity ligation strategy. The principle is based on targeting the antigenic protein with two different aptamers in order to bring the ligation DNA region into close proximity and initiate ligation-dependent qPCR(quantitative PCR) amplification (Figure 2). The protein recognition is thus translated into a detectable qPCR signal allowing sensitive quantification of coronavirus. Ct(Cycle Threshold)value changes were used to monitor the presence and concentration of the virus in serum samples. The proposed technique offers a universal diagnosis platform for coronavirus and other pathogens. Concerning therapy, the authors studied its feasibility for screening and investigation of potential neutralizing aptamers [55].
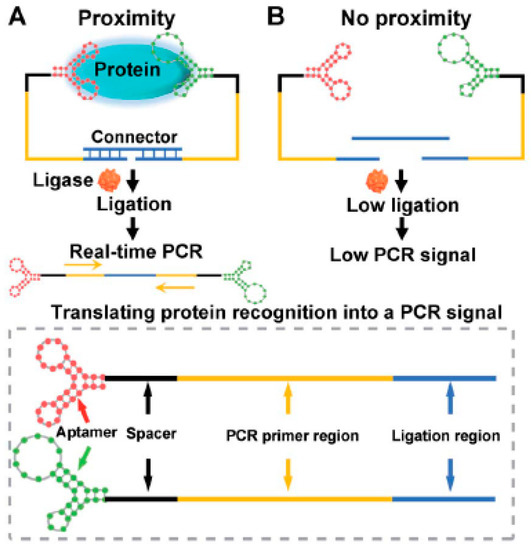
Figure 2. Schematic representation of the aptamer-assisted proximity ligation assay for COVID-19 antigens. The aptamer- proximity ligation assay has two important components: two proximity ligation probes: an aptamer region for target recognition and a spacer region to minimize the structural steric hindrance(obstruction). (A)The binding of the two aptamers to the same protein target could bring the ligation region into close proximity. Then, the two ligated aptamers hybridize with the connector template, to form a DNA (Desoxyribonucleic acid) complex used for subsequent ligation-dependent qPCR (quantitative PCR) amplification. (B)The absence of the target hampers the proximity of the two probes leading to a low ligation event and weak nonspecific qPCR signals. Reprinted with permission from [56].
Despite the excellent characteristics of aptamers, few research reports explored their advantages in COVID-19 diagnosis. Based on the excellent results of the aptasensors discussed above, more techniques will be certainly described in the future by exploring the different sequences selected either for SARS-CoV or SARS-CoV-2.
4.3. Antisense Oligonucleotides-Based Biosensing
Based on the predictable and specific hybridization of complementary bases, DNA-RNA hybridization has been widely used in RT-PCR and other biomedical diagnosis techniques. Hybridization is mainly dependent on melting temperature; complementary strands can hybridize with each other when the temperature is slightly lower than their melting temperature, while a single mismatch can cause the melting temperature to decrease significantly [56]. Qui et al. reported a complementary DNA-based biosensor for the selective detection of viral sequences including RdRp, ORF1ab, and E genes from SARS-Cov-2. In this work, dual functional biosensing was undergone by combining the plasmonic photothermal effect (PPT) and localized surface plasmon resonance (LSPR)transduction The principle is based on the hybridization between the thiol-cDNA receptors immobilized on two dimensional gold nanoisland-chip and the target genes. The PPT heat increases in the situ hybridization temperature thus allowing the discrimination of the specific gene in a multigene mixture. On the other hand, LSPR is highly sensitive to local variation, including the refractive index change and molecular binding thus allowingreal-time and label-free monitoring [57]. Electrochemical detection was also applied in the antisense-oligonucleotide-based biosensing of SARS-- genetic material. In this context, Alafeef et al developed a paper-based electrochemical platform based on a ssDNA-capped gold nanoparticles as biosensing element of SARS-CoV-2 RNA. For enhanced analytical performances, four probes were designed to bind two regions within the protein N gene. The specific cDNA (complementary DNA)-viral RNA hybridization induces charge and electron mobility changes on the graphene conductive film thus inducing a potential variation. Based on this principle, the digital electrochemical detection of SARS-CoV-2 RNA was successfully carried out in clinical samples with high sensitivity and selectivity within a short time of 5 min [58].
Owing to their simple synthesis and low cost, antisense oligonucleotides could be more advantageous than the other bioreceptors discussed in this review including aptamers and antibodies.
Table 3 and Table 4 summarize the different strategies based on nucleic acid binding applied for COVID-19 detection.
Table 3. Nucleic acid-based-biosensors for SARS-CoV-2 detection.
| Type | Detectionmethod | Target Genes | LOD | Time | Portability | Ref |
|---|---|---|---|---|---|---|
| CRISPR | All-In-One Dual CRISPR-Cas12a | SARS-CoV-2 N RNA | 4.6 copies/µL | 40 min | Nonportable | [59] |
| CRISPR-Cas12 based LFA + RT-LAMP | E and N protein genes | 10 copies/μL | <40 min | Portable | [60] | |
| CRISPR-Cas9-based, LFA | E and Orf1ab genes | 4 copies/µL | <1 h | Nonportable | [47] | |
| CRISPR/dCas9 | SARS-CoV-2 N1, N2, and N3 genes | Not available | 90 min | Portable | [61] | |
| CRISPR-Cas13 based lateral flowreadout (SHERLOCK platform) | S gene | 0.5 copies/µL | 35–70 min | Portable | [44] | |
| CRISPR-Cas13 | ORF genes | 106 copies/μL | 10 min | Nonportable | [45] | |
| CRISPR Cas12a/gRNAbased RPA | ORF1ab gene + the N proteingene | 4 × 10−1 copies/µL | ≈50 min | Nonportable | [62] | |
| AapCas12bbased LAMP (SHERLOCK platform) | N gene | 2 copies/µL | <80 min | Nonportable | [41] | |
| CRISPR-Cas13 based lateral flow | S and ORF1ab protein genes | 10 copies/µL | <1h | Nonportable | [42] | |
| CRISPR-Cas13-nCoV | ORF1ab genes | 3 copies/µL | 40 min | Nonportable | [63] | |
| RPA-CRISPR-Cas12+ lateral flowsystem | ORF1ab genes | 10 copies/μL | 10 min | Portable | [46] | |
| Antisense oligonucleotides | PPTeffect + localized SPR | RdRp, ORF1ab, and E genes | 113 × 103 copies/µL | Real time | Nonportable | [57] |
| DNAnanoswitchbasedNASBA | SARS-CoV-2 RNA | ≈100 copies/µL | 2 h | Nonportable | [64] | |
| LAMP + RAMP | ORF1ab | LAMP: 1 copies/µL RAMP: 10−1 copies/µL |
Real time | Nonportable | [65] | |
| RT-LAMP | ORF1ab + S gene | ORF1ab gene: 0.8 copies/µL S gene: 8 copies/µL |
60 min | Nonportable | [66] | |
| RT-LAMP | ORF1a/b, S and N genes | 8 × 10−2 copies/µL | 30 min | Nonportable | [67] | |
| RT-LAMP | Nsp3 gene | 6.66 copies/µL | 30 min | Nonportable | [68] | |
| RT-LAMP | nucleotide 2941-3420 ofthe COVID-19 completegenome (MN908947) | Not available | <30 min | Portable | [69] | |
| RT-LAMP | ORF1ab gene | 10 copies/µL | 15–40 min | Nonportable | [70] | |
| Graphene-basedelectrochemical+ Au-NPs | N gene | 6.9 copies/μL | <5 min | Portable | [58] | |
| Plasmoniceffectbasedcolorimetricbiosensing (Au-NPs) | N gene | 40 × 10−2 copies/µL | ≈10 min | Nonportable | [71] | |
| Isothermalrollingcircleamplification | N + S genes | 10−3 copy/µL | <2 h | Nonportable | [72] |
LFA: Lateral flow Assay, RT-LAMP (Reverse transcription loop-mediated isothermal amplification), CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats), gRNA (Guide ribonucleic acid), RPA (recombinase polymerase isothermal amplification), PPT (photothermal effect), DNA (Desoxyribonucleic acid), NASBA (Nucleic acid sequence-based amplification), RAMP (repeat-associated mysterious proteins).
Table 4. Aptamer-based-biosensors for SARS-CoV-2 detection.
| Detectionmethod | Target Genes | LOD | Time | Portability | Ref |
|---|---|---|---|---|---|
| Intrinsicsiliconthin film transistor (FET) | S protein | 0.75 ng/mL | Unknown | Nonportable | [52] |
| ELISA + colloidalgoldimmuno-chromatographicstrips | N protein | 1 ng/mL | 15 min | Nonportable | [53] |
| ELAA | N protein | 10 ng/mL | Unknown | Nonportable | [54] |
| aptamer-assistedproximityligationassay | serumnucleocapsidprotein | 37.5 ng/ mL | 2 h | Nonportable | [55] |
ELISA (Enzyme linked Immunosorbent Assay), ELAA (Enzyme Linked Aptamer Assay).
References
- Catanante, G.; Rhouati, A.; Hayat, A.; Marty, J.L. An overview of recent electrochemical immunosensing strategies for mycotoxins detection. Electroanalysis 2016, 28, 1750–1763.
- Bukkitgar, S.D.; Shetti, N.P.; Aminabhavi, T.M. Electrochemical investigations for covid-19 detection-a comparison with other viral detection methods. Chem. Eng. J. 2020, 127575, in press.
- Mohamed, M.A. Wearable miniaturized electrochemical sensors: Benefits and challenges. Electrochemistry 2018, 15, 147–185.
- Fabiani, L.; Saroglia, M.; Galatà, G.; De Santis, R.; Fillo, S.; Luca, V.; Faggioni, G.; D’Amore, N.; Regalbuto, E.; Salvatori, P.; et al. Magnetic beads combined with carbon black-based screen-printed electrodes for covid-19: A reliable and miniaturized electrochemical immunosensor for sars-cov-2 detection in saliva. Biosens. Bioelectron. 2020, 171, 112686.
- Yang, Y.; Song, Y.; Bo, X.; Min, J.; Pak, O.S.; Zhu, L.; Wang, M.; Tu, J.; Kogan, A.; Zhang, H. A laser-engraved wearable sensor for sensitive detection of uric acid and tyrosine in sweat. Nat. Biotechnol. 2020, 38, 217–224.
- Torrente-Rodríguez, R.M.; Lukas, H.; Tu, J.; Min, J.; Yang, Y.; Xu, C.; Rossiter, H.B.; Gao, W. Sars-cov-2 rapidplex: A graphene-based multiplexed telemedicine platform for rapid and low-cost covid-19 diagnosis and monitoring. Matter 2020, 3, 1981–1998.
- Eissa, S.; Zourob, M. Development of a low-cost cotton-tipped electrochemical immunosensor for the detection of sars-cov-2. Anal. Chem. 2020, 93, 1826–1833.
- Liu, J.; Chen, X.; Wang, Q.; Xiao, M.; Zhong, D.; Sun, W.; Zhang, G.; Zhang, Z. Ultrasensitive monolayer mos2 field-effect transistor based DNA sensors for screening of down syndrome. Nano Lett. 2019, 19, 1437–1444.
- Zhang, X.; Qi, Q.; Jing, Q.; Ao, S.; Zhang, Z.; Ding, M.; Wu, M.; Liu, K.; Wang, W.; Ling, Y. Electrical probing of COVID-19 spike protein receptor binding domain via a graphene field-effect transistor. arXiv 2020, in press.
- Geim, A.K.; Novoselov, K.S. The rise of graphene. In Nanoscience and Technology: A Collection of Reviews from Nature Journals; World Scientific: Singapore, 2010; pp. 11–19.
- Ward, S.; Lindsley, A.; Courter, J.; Assa’ad, A. Clinical testing for covid-19. J. Allergy Clin. Immunol. 2020, 146, 23–34.
- Marguet, S.L.; Le-Schulte, V.T.Q.; Merseburg, A.; Neu, A.; Eichler, R.; Jakovcevski, I.; Ivanov, A.; Hanganu-Opatz, I.L.; Bernard, C.; Morellini, F. Treatment during a vulnerable developmental period rescues a genetic epilepsy. Nat. Med. 2015, 21, 1436.
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G. Rapid detection of covid-19 causative virus (sars-cov-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 2020, 14, 5135–5142.
- Chen, C.; Wang, J. Optical biosensors: An exhaustive and comprehensive review. Analyst 2020, 145, 1605–1628.
- Nangare, S.N.; Patil, P.O. Engineering. Affinity-based nanoarchitectured biotransducer for sensitivity enhancement of surface plasmon resonance sensors for in vitro diagnosis: A Review. ACS Biomater. Sci. Eng. 2020, 7, 2–30.
- Das, C.M.; Guo, Y.; Yang, G.; Kang, L.; Xu, G.; Ho, H.P.; Yong, K.T. Gold nanorod assisted enhanced plasmonic detection scheme of covid-19 sars-cov-2 spike protein. Adv. Theory Simul. 2020, 3, 2000185.
- Cai, X.-F.; Chen, J.; Hu, J.; Long, Q.-X.; Deng, H.-J.; Liu, P.; Fan, K.; Liao, P.; Liu, B.-Z.; Wu, G.-C. A peptide-based magnetic chemiluminescence enzyme immunoassay for serological diagnosis of coronavirus disease 2019. J. Infect. Dis. 2020, 222, 189–193.
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W. Development and clinical application of a rapid igm-igg combined antibody test for sars-cov-2 infection diagnosis. J. Med. Virol. 2020, 92, 1518–1524.
- Jääskeläinen, A.J.; Kekäläinen, E.; Kallio-Kokko, H.; Mannonen, L.; Kortela, E.; Vapalahti, O.; Kurkela, S.; Lappalainen, M. Evaluation of commercial and automated sars-cov-2 igg and iga elisas using coronavirus disease (covid-19) patient samples. Euro Surveill. 2020, 25, 2000603.
- Huang, Z.; Chen, H.; Xue, M.; Huang, H.; Zheng, P.; Luo, W.; Liang, X.; Sun, B.; Zhong, N. Characteristics and roles of severe acute respiratory syndrome coronavirus 2-specific antibodies in patients with different severities of coronavirus 19. Clin. Exp. Immunol. 2020, 202, 210–219.
- Roda, A.; Cavalera, S.; Di Nardo, F.; Calabria, D.; Rosati, S.; Simoni, P.; Colitti, B.; Baggiani, C.; Roda, M.; Anfossi, L. Dual lateral flow optical/chemiluminescence immunosensors for the rapid detection of salivary and serum IgA in patients with COVID-19 disease. Biosens. Bioelectron. 2020, 172, 112765.
- Diao, B.; Wen, K.; Chen, J.; Liu, Y.; Yuan, Z.; Han, C.; Chen, J.; Pan, Y.; Chen, L.; Dan, Y.J.M. Diagnosis of acute respiratory syndrome coronavirus 2 infection by detection of nucleocapsid protein. medRxiv 2020, in press.
- Mahari, S.; Roberts, A.; Shahdeo, D.; Gandhi, S. Ecovsens-ultrasensitive novel in-house built printed circuit board based electrochemical device for rapid detection of ncovid-19 antigen, a spike protein domain 1 of sars-cov-2. bioRxiv 2020, in press.
- Rashed, M.Z.; Kopechek, J.A.; Priddy, M.C.; Hamorsky, K.T.; Palmer, K.E.; Mittal, N.; Valdez, J.; Flynn, J.; Williams, S.J. Rapid detection of sars-cov-2 antibodies using electrochemical impedance-based detector. Biosens. Bioelectron. 2021, 171, 112709.
- Hashemi, S.A.; Behbahan, N.G.G.; Bahrani, S.; Mousavi, S.M.; Gholami, A.; Ramakrishna, S.; Firoozsani, M.; Moghadami, M.; Lankarani, K.B.; Omidifar, N. Ultra-sensitive viral glycoprotein detection nanosystem toward accurate tracing sars-cov-2 in biological/non-biological media. Biosens. Bioelectron. 2021, 171, 112731.
- Djaileb, A.; Charron, B.; Jodaylami, M.H.; Thibault, V.; Coutu, J.; Stevenson, K.; Forest, S.; Live, L.S.; Boudreau, D.; Joelle, N.; et al. A Rapid and Quantitative Serum Test for SARS-CoV-2 Antibodies with Portable Surface Plasmon Resonance Sensing. Available online: (accessed on 4 February 2021).
- Kim, H.-Y.; Lee, J.-H.; Kim, M.J.; Park, S.C.; Choi, M.; Lee, W.; Ku, K.B.; Kim, B.T.; Park, E.C.; Kim, H.G. Development of a SARS-CoV-2-specific biosensor for antigen detection using scFv-Fc fusion proteins. Biosens. Bioelectron. 2020, 175, 112868.
- Lee, J.H.; Choi, M.; Jung, Y.; Lee, S.K.; Lee, C.S.; Kim, J.; Kim, J.; Kim, N.H.; Kim, B.T.; Kim, H.G. A novel rapid detection for sars-cov-2 spike 1 antigens using human angiotensin converting enzyme 2 (ace2). Biosens. Bioelectron. 2021, 171, 112715.
- Huang, C.; Wen, T.; Shi, F.J.; Zeng, X.Y.; Jiao, Y.J. Rapid detection of igm antibodies against the sars-cov-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega 2020, 5, 12550–12556.
- Liu, H.; Dai, E.; Xiao, R.; Zhou, Z.; Zhang, M.; Bai, Z.; Shao, Y.; Qi, K.; Tu, J.; Wang, C.; et al. Development of a sers-based lateral flow immunoassay for rapid and ultra-sensitive detection of anti-sars-cov-2 igm/igg in clinical samples. Sens. Actuators B Chem. 2020, 3209, 129196.
- Xiang, J.; Yan, M.; Li, H.; Liu, T.; Lin, C.; Huang, S.; Shen, C. Evaluation of enzyme-linked immunoassay and colloidal gold-immunochromatographic assay kit for detection of novel coronavirus (sars-cov-2) causing an outbreak of pneumonia (covid-19). MedRxiv 2020, in press.
- Cady, N.C.; Tokranova, N.; Minor, A.; Nikvand, N.; Strle, K.; Lee, W.T.; Page, W.; Guignon, E.; Pilar, A.; Gibson, G.N. Multiplexed detection and quantification of human antibody response to covid-19 infection using a plasmon enhanced biosensor platform. Biosens. Bioelectron. 2021, 171, 112679.
- Pan, Y.; Li, X.; Yang, G.; Fan, J.; Tang, Y.; Zhao, J.; Long, X.; Guo, S.; Zhao, Z.; Liu, Y. Serological immunochromatographic approach in diagnosis with sars-cov-2 infected covid-19 patients. J. Infect. 2020, 81, e28–e32.
- Huang, L.; Ding, L.; Zhou, J.; Chen, S.; Chen, F.; Zhao, C.; Xu, J.; Hu, W.; Ji, J.; Xu, H.; et al. One-step rapid quantification of sars-cov-2 virus particles via low-cost nanoplasmonic sensors in generic microplate reader and point-of-care device. Biosens. Bioelectron. 2021, 171, 112685.
- Lin, Q.; Wen, D.; Wu, J.; Liu, L.; Wu, W.; Fang, X.; Kong, J. Microfluidic immunoassays for sensitive and simultaneous detection of igg/igm/antigen of sars-cov-2 within 15 min. Anal. Chem. 2020, 92, 9454–9458.
- Liu, W.; Liu, L.; Kou, G.; Zheng, Y.; Ding, Y.; Ni, W.; Wang, Q.; Tan, L.; Wu, W.; Tang, S. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against sars-cov-2. J. Clin. Microbiol. 2020, 58, e00461-20.
- Zhu, X.; Wang, X.; Han, L.; Chen, T.; Wang, L.; Li, H.; Li, S.; He, L.; Fu, X.; Chen, S.; et al. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of covid-19. Biosens. Bioelectron. 2020, 166, 112437.
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C. Nucleic acid detection with crispr-cas13a/c2c2. Science 2017, 356, 438–442.
- O’Connell, M.R. Molecular mechanisms of rna targeting by cas13-containing type vi crispr–cas systems. Mol. Biol. 2019, 431, 66–87.
- Vatankhah, M.; Azizi, A.; Sanajouyan Langeroudi, A.; Ataei Azimi, S.; Khorsand, I.; Kerachian, M.A.; Motaei, J. Crispr-based biosensing systems: A way to rapidly diagnose covid-19. Rev. Clin. Lab. Sci. 2020, 1–27.
- Joung, J.; Ladha, A.; Saito, M.; Segel, M.; Bruneau, R.; Huang, M.-l.W.; Kim, N.-G.; Yu, X.; Li, J.; Walker, B.D.J.M. Point-of-care testing for covid-19 using sherlock diagnostics. medRxiv 2020, in press.
- Zhang, F.; Abudayyeh, O.O.; Gootenberg, J.S. A protocol for detection of COVID-19 using CRISPR diagnostics. A protocol for detection of COVID-19 using CRISPR diagnostics. medRxiv 2020, in press.
- Sherlock CRISPR SARS-CoV-2 Kit; SherlockBioSciences, Inc. Available online: (accessed on 4 February 2021).
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1–10.
- Arizti-Sanz, J.; Freije, C.A.; Stanton, A.C.; Petros, B.A.; Boehm, C.K.; Siddiqui, S.; Shaw, B.M.; Adams, G.; Kosoko-Thoroddsen, T.-S.F.; Kemball, M.E. Streamlined inactivation, amplification, and cas13-based detection of sars-cov-2. Nat. Commun. 2020, 11, 1–9.
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Jessica, A.; Streithorst, J.A.; Granados, A.; et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 1–5.
- Huang, Z.; Tian, D.; Liu, Y.; Lin, Z.; Lyon, C.J.; Lai, W.; Fusco, D.; Drouin, A.; Yin, X.; Hu, T.; et al. Ultra-sensitive and high-throughput CRISPR-Powered COVID-19 diagnosis. Biosens. Bioelectron. 2020, 164, 112316.
- Lucia, C.; Federico, P.-B.; Alejandra, G.C. An ultrasensitive, rapid, and portable coronavirus sars-cov-2 sequence detection method based on crispr-cas12. bioRxiv 2020, in press.
- Xiong, E.; Jiang, L.; Tian, T.; Hu, M.; Yue, H.; Huang, M.; Lin, W.; Jiang, Y.; Zhu, D.; Zhou, X.J.A.C. Simultaneous Dual-Gene Diagnosis of sars-cov-2 Based on crispr/cas9-mediated Lateral Flow Assay. Available online: (accessed on 4 February 2021).
- Brody, E.N.; Gold, L. Aptamers as therapeutic and diagnostic agents. Rev. Mol. Biotechnol. 2000, 74, 5–13.
- Toh, S.Y.; Citartan, M.; Gopinath, S.C.B.; Tang, T.-H. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens. Bioelectron. 2015, 64, 392–403.
- Farrow, T.; Laumier, S.; Hall, S.; Sandall, I.; Hv, Z. Feasibility of a silicon thin film transistor-based aptamer sensor for COVID-19 detection. IEEE Sens. 2020, in press.
- Zhang, L.; Fang, X.; Liu, X.; Ou, H.; Zhang, H.; Wang, J.; Li, Q.; Cheng, H.; Zhang, W.; Luo, Z. Discovery of sandwich type covid-19 nucleocapsid protein DNA aptamers. Chem. Commun. 2020, 56, 10235–10238.
- Chen, Z.; Wu, Q.; Chen, J.; Ni, X.; Dai, J. A DNA aptamer based method for detection of sars-cov-2 nucleocapsid protein. VirologicaSinica 2020, 35, 351–354.
- Liu, R.; He, L.; Hu, Y.; Luo, Z.; Zhang, J. A serological aptamer-assisted proximity ligation assay for covid-19 diagnosis and seeking neutralizing aptamers. Chem. Sci. 2020, 11, 12157–12164.
- Sefah, K.; Phillips, J.A.; Xiong, X.; Meng, L.; Van Simaeys, D.; Chen, H.; Martin, J.; Tan, W. Nucleic acid aptamers for biosensors and bio-analytical applications. Analyst 2009, 134, 1765–1775.
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano 2020, 14, 5268–5277.
- Song, Y.; Song, J.; Wei, X.; Huang, M.; Sun, M.; Zhu, L.; Lin, B.; Shen, H.; Zhu, Z.; Yang, C. Discovery of aptamers targeting the receptor-binding domain of the sars-cov-2 spike glycoprotein. Anal. Chem. 2020, 92, 9895–9900.
- Harris, N.C.; Kiang, C.-H. Defects can increase the melting temperature of DNA−nanoparticle assemblies. J. Phys. Chem. B 2006, 110, 16393–16396.
- Alafeef, M.; Dighe, K.; Moitra, P.; Pan, D. Rapid, ultrasensitive, and quantitative detection of sars-cov-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano 2020, 14, 17028–17045.
- Ding, X.; Yin, K.; Li, Z.; Liu, C. All-in-one dual crispr-cas12a (aiod-crispr) assay: A Rapid, Ultrasensitive, and Quantitative Detection of SARS-CoV-2 Using Antisense Oligonucleotides Directed Electrochemical Biosensor Chip. ACS Nano 2020, 11, 4711.
- Moon, J.; Kwon, H.-J.; Yong, D.; Lee, I.-C.; Kim, H.; Kang, H.; Lim, E.-K.; Lee, K.-S.; Jung, J.; Park, H.G.; et al. Colorimetric detection of sars-cov-2 and drug-resistant ph1n1 using crispr/dcas9. ACS Sens. 2020, 5, 4017–4026.
- Hou, T.; Zeng, W.; Yang, M.; Chen, W.; Ren, L.; Ai, J.; Wu, J.; Liao, Y.; Gou, X.; Li, Y. Development and Evaluation of a Crispr-Based Diagnostic for 2019-Novel Coronavirus. Available online: (accessed on 4 February 2021).
- Zhou, L.; Chandrasekaran, A.R.; Punnoose, J.A.; Bonenfant, G.; Charles, S.; Levchenko, O.; Badu, P.; Cavaliere, C.; Pager, C.T.; Halvorsen, K. Programmable low-cost DNA-based platform for viral RNA detection. bioRxiv 2020, 6, eabc6246.
- El-Tholoth, M.; Bau, H.H.; Song, J. A Single and Two-Stage, Closed-Tube, Molecular Test for the 2019 Novel Coronavirus (COVID-19) at Home, Clinic, and Points of Entry. ChemRxiv 2020, in press.
- Yan, C.; Cui, J.; Huang, L.; Du, B.; Chen, L.; Xue, G.; Li, S.; Zhang, W.; Zhao, L.; Sun, Y. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020, 26, 773–779.
- Huang, W.E.; Lim, B.; Hsu, C.C.; Xiong, D.; Wu, W.; Yu, Y.; Jia, H.; Wang, Y.; Zeng, Y.; Ji, M. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020, 13, 950–961.
- Park, G.-S.; Ku, K.; Baek, S.-H.; Kim, S.-J.; Kim, S.I.; Kim, B.-T.; Maeng, J.-S. Development of reverse transcription loop-mediated isothermal amplification (RT-LAMP) assays targeting SARS-CoV-2. J. Mol. Diagn. 2020, 22, 729–735.
- Lamb, L.E.; Bartolone, S.N.; Ward, E.; Chancellor, M.B. Rapid Detection of Novel Coronavirus (COVID19) by Reverse Transcription-Loop-Mediated Isothermal Amplification. Available online: (accessed on 4 February 2021).
- Yu, L.; Wu, S.; Hao, X.; Li, X.; Liu, X.; Ye, S.; Han, H.; Dong, X.; Li, X.; Li, J. Rapid colorimetric detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform: iLACO. Clin. Chem. 2020, 66, 975–977.
- Moitra, P.; Alafeef, M.; Dighe, K.; Frieman, M.; Pan, D. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14, 7617–7627.
- Chaibun, T.; Puenpa, J.; Ngamdee, T.; Boonapatcharoen, N.; Athamanolap, P.; O’Mullane, A.P.; Vongpunsawad, S.; Poovorawan, Y.; Yin, L.S.; Lertanantawong, B. Rapid Electrochemical Detection of Coronavirus SARS-CoV-2. Nat. Commun. 2020, 12, 1–10.





