
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Oscar Campuzano | + 2872 word(s) | 2872 | 2021-03-24 09:09:41 | | | |
| 2 | Lily Guo | + 641 word(s) | 3513 | 2021-03-29 09:53:28 | | | | |
| 3 | Lily Guo | + 641 word(s) | 3513 | 2021-03-29 09:54:04 | | |
Video Upload Options
The RBM20 gene encodes the muscle-specific splicing factor RNA-binding motif 20, a regulator of heart-specific alternative splicing. Nearly 40 potentially deleterious variants in RBM20 have been reported in the last ten years, being found to be associated with highly arrhythmogenic events in familial dilated cardiomyopathy. Frequently, malignant arrhythmias can be a primary manifestation of disease. The early recognition of arrhythmic genotypes is crucial in avoiding lethal episodes, as it may have an impact on the adoption of personalized preventive measures.
1. Introduction
The RBM20 gene (Gene ID: 282996; HGNC: 27424; OMIM: 613171; Gencode Gene: ENSG00000203867.7) is located on the long arm of chromosome 10 at position 25.2 (10q25.2) and it encodes the RNA binding motif protein-20 (RBM20). This gene comprises 14 exons (UniProtKB: Q5T481; RefSeq: NM_001134363; Gencode Transcript: ENST00000369519.3) that encode three conserved functional domains: two zinc finger (ZnF) domains and one RNA recognition motif (RRM)-type RNA-binding domain. In addition, sequence alignment from various vertebrate species shows three other conserved regions: a leucine (L)-rich region at the N-terminus, an arginine/serine (RS)-rich region just downstream from the RRM domain, and a glutamate (E)-rich region between the RS-rich region and ZnF2 domain [1] (Figure 1). The phosphorylation of arginine–serine–arginine–serine–proline residues in the RS region (RSRSP stretch) is necessary for RBM20 nuclear localization [2].
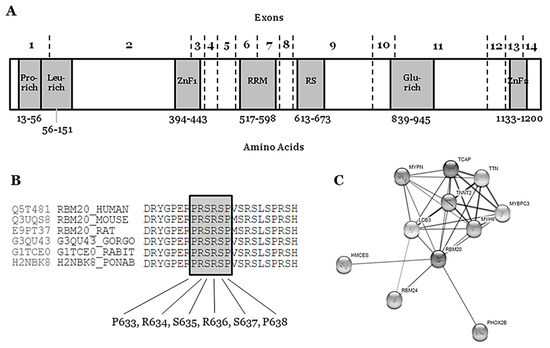
Figure 1. Structure and network of the RBM20 protein. (A) Glu-rich: Glutamate rich region; Leu-rich: Leucine rich region; Pro-rich: Proline rich region; RRM: RNA Recognition Motif; RS: Arginine-Serine Domain; ZnF1: Zinc Finger region 1; ZnF2: Zinc Finger region 2. (B) Conservation between species of RS region (amino acids 634–638). (C) Network of ten closest proteins to RBM20.
The RBM20 gene is highly expressed during human fetal development (mainly 11–20 weeks of gestation) and in heart and skeletal muscle [3]. The protein (length: 1227 amino acids; mass: 134,357 Da) binds RNA and regulates the splicing of a subset of genes that are involved in cardiac development [4]. It is one of the few heart-specific splicing factors that regulate alternative splicing events of many genes, including TTN and LDB3 [5][6], and it is associated with sarcomere assembly, ion transport, and diastolic function [7], as well as the expression of calcium handling, rendering high arrhythmic risk to RBM20 carrier patients [8] (Figure 1).
In 2012, Gu et al., reported an animal model with a deficient titin splicing identifying a loss-of-function mutation in RBM20 as the underlying cause for the pathological titin isoform expression. The affected rats had a 95 kb deletion that removed exons 2–14. The missing exons encode the RNA binding motif-, the RS-, and the Zn2+ finger domains. They determined that RBM20 is required for titin splicing, as well as for the alternative splicing of many other conserved cardiac genes, as revealed by deep sequencing of rat and human samples. Defective splicing that is caused by the RBM20 mutation in rats resulted in features resembling those of humans carrying RBM20 mutations, including left ventricular dilatation, subendocardial fibrosis, arrhythmia, and sudden death. Adenoviral gene delivery to re-express RBM20 in deficient cardiomyocytes was performed and reconstituted expression of the short titin isoform. They investigated an individual carrying a heterozygote p.Ser635Ala mutation in RBM20 to validate RBM20 dependent titin splicing and its relevance for human disease. On the protein level, the heterozygote RBM20 mutation p.Ser635Ala shifted human titin isoform expression with an increased molecular weight that was similar to the larger isoform expressed in heterozygous rats [7].
2. Rare RBM20 Variants in Familial Dilated Cardiomyopathy
Nowadays, individuals carrying RBM20 pathogenic variants are at a high risk of AR-DCM and early ICD implantation should be discussed [9]. We performed an exhaustive review of the literature concerning RBM20 and DCM published before October 2020. The data were collected from: HGMD (www.hgmd.org), ClinVar (www.ncbi.nlm.nih.gov/clinvar/intro), National Center for Biotechnology Information SNP database (www.ncbi.nlm.nih.gov/SNP), Index Copernicus (www.en.indexcopernicus.com), Google Scholar (scholar.google.es), Springer Link (www.link.springer.com), Science Direct (www.sciencedirect.com), Excerpta Medica Database (www.elsevier.com/solutions/embase-biomedical-research), and IEEE Xplore Digital Library (www.ieeexplore.ieee.org/Xplore/home.jsp). Genetic variants that were identified in articles were contrasted with variant data from Exome Variant Server (EVS, www.evs.gs.washington.edu/EVS) and Genome Aggregation Database (gnomAD, www.gnomad.broadinstitute.org), including recently added data regarding copy number variation (CNV). In addition, we consulted data for amino acid sequence and conservation between species in UniProt (www.uniprot.org). The variants were classified according to the American College of Medical Genetics and Genomics/Association for Molecular Pathology (ACMG/AMP) standards and guidelines for the interpretation of sequence variants [10] and described using the HGVS recommendations for the description of sequence variants [11][12]. Concerning frequency of disease-causing variants that are associated with rare inherited diseases, the vast majority of deleterious variants are extremely rare (<0.01%) [13]. ClinGen (www.clinicalgenome.org/), CardioClassifier (www.cardioclassifier.org), CardioBoost (www.cardiodb.org/cardioboost/), and VarSome (www.varsome.com) were consulted. Finally, all of the investigators discussed data and came to a consensus on the final classification of variants to avoid any bias.
In the RBM20 gene, thirty-six rare non-synonymous variants have been reported as causes of FDCM. Of these, thirty-four are mapped within exons and two in intronic zones (c.1880 + 4_1880 + 6dupAGG and c.1528-1G>C -CI1516347 and CS183215 respectively-). Most of the exonic variants (32/34) are missense variants, and two are nonsense variants [p.(Arg688*) and p.(Gly1031*)—CM1516720 and CM1111136, respectively]. Importantly, all of the rare variants are reported in heterozygosis status. After comprehensive analysis of all clinical and genetic data published so far, five rare variants should be considered, likely benign (LB), eighteen of ambiguous role or variants of uncertain significance (VUS), nine as likely pathogenic (LP), and four as pathogenic (P) for FDCM (Table 1).
Table 1. Data of variants in RBM20 potentially associated with Dilated Cardiomyopathy.
| Nucleotide Change | Protein Change | dbSNP | gnomAD (MAF%) | ClinVar (Disease) | HGMD (Disease) | CC | ACMG Score | RBM20 Domain | Arrhythmogenic Phenotype |
|---|---|---|---|---|---|---|---|---|---|
| c.247C > A | p.(Leu83Ile) | rs536357058 | 1/155140 (0.0006%) | VUS (DCM) | CM1111132 | VUS | VUS | Exon 2 | Yes |
| (DM; DCM) | Leucine-rich region | ||||||||
| c.680G > T | p.(Gly227Val) | rs202238753 | 225/185204 (0.12%) | LB (DCM) | CM1821953 | LB | VUS | Exon 2 | No |
| (DM; DCM) | |||||||||
| c.769A > G | p.(Thr257Ala) | rs1418674149 | 1/153900 (0.0006%) | NA | CM1815813 | VUS | VUS | Exon 2 | Yes |
| (DM; DCM) | |||||||||
| c.1175G > A | p.(Arg392Gln) | rs751788298 | 3/185862 (0.0016%) | NA | CM1815814 | VUS | VUS | Exon 2 | NA |
| (DM; DCM) | |||||||||
| c.1364C > T | p.(Ser455Leu) | rs189569984 | 862/153884 (0.56%) | LB | NA | LB | LB | Exon 4 | No |
| c.1494C > A | p.(Ser498Arg) | rs774916799 | 2/153882 (0.0013%) | VUS (DCM) | CM1815816 | VUS | VUS | Exon 4 | Yes |
| (DM; DCM) | |||||||||
| c.1528-1G > C | - | rs534513476 | NA | NA | CS183215 | VUS | P | Intron 5–6 | Yes |
| (DM; DCM) | |||||||||
| c.1603G > A | p.(Val535Ile) | rs183007628 | 6/188686 (0.0031%) | VUS (DCM) | CM107458 | VUS | VUS | Exon 6 | Yes |
| (DM; DCM) | RNA Recognition Motif | ||||||||
| c.1760T > A | p.(Leu587His) | NA | NA | NA | CM1815817 | VUS | VUS | Exon 7 | Yes |
| (DM; DCM) | RNA Recognition Motif | ||||||||
| c.1764T > G | p.(Ile588Met) | NA | NA | NA | CM183216 | VUS | VUS | Exon 7 | Yes |
| (DM; DCM) | RNA Recognition Motif | ||||||||
| c.1880 + 4_1880 + 6dupAGG | - | rs1227694990 | 200/187706 (0.1%) | LB (DCM) | CI1516347 | VUS | VUS | Intron 7−8 | No |
| (DM; DCM) | |||||||||
| c.1898C > T | p.(Pro633Leu) | rs747880281 | 1/151498 (0.0006%) | VUS (DCM) | NA | VUS | P | Exon 9 | Yes |
| Arginine-Serine Domain | |||||||||
| c.1900C > T | p.(Arg634Trp) | NA | NA | NA | CM107456 | VUS | LP | Exon 9 | Yes |
| (DM; DCM) | Arginine-Serine Domain | ||||||||
| c.1901G > A | p.(Arg634Gln) | rs267607001 | 1/152378 (0.0006%) | P (DCM) | CM095004 | VUS | LP | Exon 9 | Yes |
| c.1901G > T | p.(Arg634Leu) | (DM; DCM) | Arginine-Serine Domain | ||||||
| c.1903T > G | p.(Ser635Ala) | NA | NA | NA | CM125867 | VUS | LP | Exon 9 | Yes |
| (DM; DCM) | Arginine-Serine Domain | ||||||||
| c.1906C > A | p.(Arg636Ser) | rs267607002 | NA | NA | CM095005 | LP | LP | Exon 9 | Yes |
| (DM; DCM) | Arginine-Serine Domain | ||||||||
| c.1906C > T | p.(Arg636Cys) | rs267607002 | NA | NA | CM107457 | LP | LP | Exon 9 | Yes |
| (DM; DCM) | Arginine-Serine Domain | ||||||||
| c.1907G > A | p.(Arg636His) | rs267607004 | NA | NA | CM095006 | VUS | LP | Exon 9 | Yes |
| (DM; DCM) | Arginine-Serine Domain | ||||||||
| c.1909A > G | p.(Ser637Gly) | rs267607005 | NA | NA | CM095007 | VUS | LP | Exon 9 | Yes |
| (DM; DCM) | Arginine-Serine Domain | ||||||||
| c.1913C > T | p.(Pro638Leu) | rs267607003 | NA | NA | CM095008 | VUS | LP | Exon 9 | Yes |
| (DM; DCM) | Arginine-Serine Domain | ||||||||
| c.1997G > A | p.(Arg666Gln) | rs202011408 | 5/154830 (0.003%) | NA | CM1716804 | VUS | VUS | Exon 9 | Yes |
| (DM; DCM) | Arginine-Serine Domain | ||||||||
| c.2021A > G | p.(Asp674Gly) | rs1475557145 | 1/155286 (0.0006%) | VUS (DCM) | NA | VUS | VUS | Exon 9 | NA |
| c.2042A > G | p.(Tyr681Cys) | rs372048968 | 23/186630 (0.01%) | VUS (DCM) | CM1815818 | LB | LB | Exon 9 | No |
| (DM; DCM) | |||||||||
| c.2062C > T | p.(Arg688Ter) | rs794729150 | 1/31344 (0.003%) | VUS (DCM) | CM1516720 | VUS | P | Exon 9 | Yes |
| (DM; DCM) | |||||||||
| c.2109G > C | p.(Arg703Ser) | rs988797559 | 2/186026 (0.001%) | NA | CM1111134 | VUS | VUS | Exon 9 | Yes |
| (DM; DCM) | |||||||||
| c.2147G > A | p.(Arg716Gln) | rs375798246 | 21/155108 (0.013%) | VUS (DCM) | NA | LB | LB | Exon 9 | No |
| c.2282G > A | p.(Arg761Gln) | rs556897484 | 4/156496 (0.002%) | NA | NA | VUS | VUS | Exon 9 | NA |
| c.2662G > A | p.(Asp888Asn) | rs201370621 | 603/155726 (0.3%) | VUS (DCM) | NA | LB | LB | Exon 11 | No |
| c.2737G > A | p.(Glu913Lys) | rs397516607 | NA | LP (DCM) | NA | LP | LP | Exon 11 | Yes |
| c.2741T > C | p.(Val914Ala) | rs794729154 | NA | NA | NA | VUS | VUS | Exon 11 | Yes |
| c.2714T > A | p.(Met950Lys) | NA | NA | NA | NA | VUS | VUS | Exon 11 | NA |
| c.3091G > T | p.(Gly1031Ter) | rs794729157 | NA | NA | CM1111136 | VUS | P | Exon 11 | Yes |
| (DM; DCM) | |||||||||
| c.3115C > T | p.(Pro1039Ser) | rs727503392 | 40/188260 (0.02%) | LB (DCM) | CM1815819 | VUS | LB | Exon 11 | No |
| (DM; DCM) | |||||||||
| c.3242C > G | p.(Pro1081Arg) | rs1268330519 | NA | NA | CM1111137 | VUS | VUS | Exon 12 | Yes |
| (DM; DCM) | |||||||||
| c.3545G > A | p.(Arg1182His) | rs563762318 | 47/185298 (0.025%) | LB (DCM) | CM1510988 | VUS | VUS | Exon 12 | Yes |
| (DM; DCM) | Zinc Finger domain 2 | ||||||||
| c.3616G > A | p.(Glu1206Lys) | rs757389650 | 8/181254 (0.004%) | VUS (DCM) | CM1111138 | VUS | VUS | Exon 14 | NA |
Rare variants that are definitely classified as LB can be discarded as causal for FDCM, mainly due to high frequency in the population. However, we cannot discard their potential role as phenotype modifiers. No VUS can be discarded as a potential cause of FDCM—a variant currently classified as VUS means that conclusive data do not exist, so additional studies are needed in order to clarify the definite role in FDCM. These 18 rare non-synonymous VUS in RBM20 should be interpreted with caution by a group of experts, as clinical translation should be personalized, accounting for not only all published data, but also family segregation and phenotype of each patient [14]. The four rare variants classified as P are located in intron 5–6—c.1528-1G>C/IVS5asG>C-1—the end of exon 9—p.(Pro633Leu), p.(Arg688*)—and exon 11—p.(Gly1031*). These variants are considered definitely P due to their extremely low frequency in global population, in silico predictions and functional studies. Most of the variants classified as LP are located in exon 9 (RS domain, amino acids 634–638), suggesting a hot-spot for malignant arrhythmias in FDCM. Actually, a recent study identified two RBM20 regions (exons 9 and 11) with a significant risk for cardiomyopathy, ventricular and atrial arrhythmias, and even SCD [15]. We only identified one rare variant classified as LP and located out of this hot-spot (p.Glu913Lys, Glutamic acid-rich domain). None of these LP variants can be classified as definitely P for FDCM mainly due to a lack of functional data. However, their highly malignant role is supported by low frequencies in global population databases as well as a conserved domain between species.The hot-spot P-R-S-R-S-P between p.(Pro633) and p.(Pro638) contains crucial amino acids for the protein structure and function (Figure 1) (Table 1). In consequence, any amino acid modification inside this zone implies, a priori, a high probability of damaging effect in the protein structure and function. In the first amino acid of this hot-spot, only one rare variant has been recently reported—p.(Pro633Leu). Clinical, genetic, and functional studies confirmed the pathogenic role of this rare variant [16]. Importantly, in this recent study, the authors also suggested that the upregulation of RBM20 may be a viable therapeutic strategy for RBM20-related DCM. In the amino acid 634, two changes have been reported as LP in FDCM [p.(Arg634Trp) and p.(Arg634Gln), CM107456 and CM095004, respectively] [2][17]. In p.(Ser635), only one variation is reported—p.(Ser635Ala), CM125867 [7]. In p.(Arg636), three changes have been published as LP in FDCM—p.(Arg636Ser), p.(Arg636Cys), and p.(Arg636His) [17][18][19][20]. In the last two amino acids, p.(Ser637) and p.(Pro638), a change has been identified in each—p.(Ser637Gly) and p.(Pro638Leu) [21][22][23][24] (Figure 1) (Table 1). The establishment of hiPSC-CMs shows that pathogenic alterations in some of these amino acids may disorganize the sarcomeric complex [8][25][26]. All these variants were identified in families including aggressive arrhythmogenic phenotypes, occasionally in individuals with discrete structural heart alterations, and with a high penetrance of the disease. Despite this fact, we cannot discard that future studies and additional evidence may allow for the identification of other rare variants that are located in different regions of RBM20 and still not associated with arrhythmogenic phenotypes in FDCM. Finally, with regard to CNVs as being potentially responsible for FDCM, to date no structural alteration has been identified in RBM20. Only two CNVs have been associated with FDCM, one located in LMNA [27] and the other in BAG3 [28], explaining <5% of FDCM cases together [29][30].The first pathogenic variant in the RBM20 gene was reported in 2009 as a novel cause for familial DCM [17]. In this first cross-sectional study, pathogenic variants in RBM20 were present in the DCM families showing high penetrance, a tent to young age at diagnosis, a notable presence of end-stage heart failure, and high mortality, according to the available information in the included individuals [18]. Nowadays, nearly 30 rare variants in RBM20 have been reported, explaining 2–3% of DCM [31][32] and supporting an aggressive arrhythmogenic phenotype with a higher risk of SCD [15][19][22][33][34] (Table 2).
Table 2. Rare variants in RBM20.
| Brauch et al., 2009 (n = 39, DCM) NC | Li et al., 2010 (n = 16, DCM) NC | Refaat et al., 2012 (n = 8, DCM) | Wells et al., 2013 (n = 19 carriers) NC | Van den Hoogenhof et al., 2018 (n = 18, DCM) | Hey et al., 2019 (n = 53, DCM) | Parikh et al., 2019 (n = 74, carriers) | |
|---|---|---|---|---|---|---|---|
| Age diagnosis | 36 ± 13.2 | 37.6 ± 9 | - | 33.8 ± 11.5 | 42 ± 14 | 37 ± 15 & | 37 ± 15 † |
| Males | 19 (49%) | 8 (50%) | 4 (50%) | 14 (82%) | 8 (44%) | 31 (58%) | - |
| Follow-up (months) | 60 (12−204) | - | 27.4 ± 15.7 | - | 71 ± 65 | 86 (24−150) | - |
| Mean LVEF | 35.3 ± 11.5 | 29.3 ± 8.6 | - | 48.8 ± 13 | 37 ± 17 | 32 ± 12 && | 40 ± 17 |
| FH SCD | 39 (100%) | - | - | - | 13 (72%) | - | 22/43 (51%) †† |
| NSVT | - | 1 (6%) | - | - | 5 (28%) | - | 21/59 (36%) |
| Sustained VT or VF | 9 (23%) | 1 (6%) | 0 | - | 8 (44%) ¶ | 11 (21%) &&& | - |
| ICD therapy | - | 0 | 1 (12.5%) | - | - | - | 9/32 (28%) ††† |
| SCD | 3 (7.7%) | 1 (6%) | 0 | - | - | 6 (11.3%) &&& | 5/60 (8%)-SCA ††† |
| AF | 3 (7.7%) | 2 (12.5%) | 3 (37.5%) * | 6 (33%) | - | 10/58 (17%) †††† | |
| HTx | 4 (mean age 28.5) | 2 (12.5%) | 1 (12.5%) | 1 (5.2%, 17 years old) + | - | 11 (21%) &&&& | 5/74 (7%) NC |
| Death | 11 (28%, mean age 45): 4 HF (mean age 54.7), 3 SCD (mean age 39) | 3 (11.5%) | 0 | 11 (57.9%) + |
- | 2 (4%, end-stage HF at 54 and 73 years old) | 3/74 (4%) NC |
Reefat et al., studied 283 individuals with DCM from the GRADE cohort (that studies the genetic background of patients carrying an ICD), including only non-transplanted patients and with no heart assist device [22]. The patients were screened for RBM20 pathogenic alterations. The mean age of subjects with DCM was 58 ± 13 years, 64% were males, and the mean follow-up time was 24.2 ± 17.1 months after ICD placement. Pathogenic alterations in RBM20 were identified in eight subjects with DCM (2.8%). Carriers of these pathogenic rare variants had a similar survival, transplantation rate, and frequency of ICD therapy as compared with non-variant carriers. Three of eight subjects carrying RBM20 alterations (37.5%) had atrial fibrillation (AF), whereas 19 subjects without rare pathogenic alterations (7.4%) had AF (p = 0.02) (Table 2). Among all of the GRADE subjects, rs35141404, a common genetic polymorphism situated in the RBM20 gene, was associated with AF (p = 0.006), and this association remained in the subset of GRADE subjects with DCM (p = 0.047) [22]. This locus have been validated by large GWAS AF studies in general population [35][36]. Haas et al., investigated gene groups in DCM patients to identify genotype-phenotype correlations. In a cohort of 639 patients, the presence of P, LP or VUS RBM20 variants identified in 15 patients provided an OR of 5.65 (1.89–16.86; p = 0.002) for ICD carrier status in DCM [31]. Van den Hoogenhof et al., compared 18 DCM patients carrying RBM20 alterations to 22 DCM patients carrying TTN alterations, and found that 44% of patients carrying any RBM20 alteration had sustained ventricular arrhythmias (Sustained VT or VF) when compared to 5% of patients carrying any TTN alteration, despite similar LVEF. No differences in non-sustained VT and AF prevalence were detected [34] (Table 2). In another multicenter registry, 72 DCM patients carrying alterations in the RBM20 gene were compared to idiopathic DCM (n = 633), TTNtv related to DCM, and LMNA-related DCM. There was a considerable family history of SCD (51%) similar to LMNA patients (44%, NS) and greater than idiopathic DCM and TTNtv DCM patients (15% in both, p < 0.001 respectevily). The carriers of RBM20 alterations were more likely to have sustained VT (25%) than idiopathic DCM cohort (2%, p < 0.001) and TTNtv variants cohort (1%, p < 0.001) and, similar to LMNA patients (21%, NS), defined as sustained VT or VF on monitoring for idiopathic DCM, TTNtv, and LMNA, and as sudden cardiac arrest or ICD discharge for RBM20 [15] (Table 2). In the largest cohort of RBM20 patients published to date, Hey et al., included a total of 80 individuals from 15 families carrying pathogenic alterations (p.Arg634Gln, p.Arg636His, p.Arg636Ser, p.Pro638Leu, and p.Glu913Lys) (Table 1 and Table 2). Ten index-patients were shown to carry the same p.Arg636Ser and seven of those shared the haplotype analyses, which suggested a common founder. The penetrance was 66% (53/80) and age-dependent. The males were both significantly younger and had lower ejection fraction at diagnosis than females (age, 29 ± 11 versus 48 ± 12 years; p < 0.01; ejection fraction, 29 ± 13% versus 38 ± 9%; p < 0.01). Subsequently, 11 of 31 affected males needed a cardiac transplant while none of 22 the affected females required this treatment (p < 0.001). However, two females died of end-stage heart failure at the age of 54 and 72, while none of the males did. Remarkably, seven young males and no females developed DCM in their teens, requiring heart transplant in four individuals before the age of 19 (p.Arg636His, p.Glu913Lys, p.639Leu). Thirty percent of RBM20-carriers with DCM (16/53) died suddenly or experienced severe ventricular arrhythmia: six died suddenly, three were successfully resuscitated from a cardiac arrest because of VF, two had episodes of sustained VT requiring cardioversion, and five received appropriated therapies by a prophylactic ICD. From the six patients that died suddenly, 1 SCD occurred in a patient with a LVEF of 30% and 5/6 SCD patients were diagnosed of DCM by post-mortem autopsy, belonging four of these cases to one of the two families carrying the same RBM20 p.Arg636Ser variant. The only available pathology report described a dilated LV and a weight of 475 g (0.8% of her body weight). For the eleven patients experiencing ventricular arrhythmia with available echocardiography, the median LVEF was median 30%; range, 10–47%. However, 36% (4/11) had a LVEF >30%. No adverse events were identified among healthy RBM20-carriers with a normal cardiac investigation. The event-free survival of male RBM20-carriers was significantly shorter when compared with female carriers (p < 0.001). [33] (Table 2).
Taking all of the data into account, RBM20 pathogenic alterations may cause disturbing cardiac contraction and impair cardiac conduction [18]. The mean age at diagnosis is around the forth decade. LVEF is usually impaired, leading to mild to severe DCM, which can lead to heart failure and eventually heart transplantation, which can be required at very young ages. In the published series, no fatal events have been detected in patients without LV structural abnormalities. However, some of these patients have been diagnosed post-mortem, as SCD was the first manifestation of the disease. Similarly to LMNA rare variants, the disease expression seems to be gender-specific. Most of RBM20 carrier’s males are diagnosed at younger ages and suffer major cardiac events before 40 years old when comparing to females (less than 5%) [33]. Lower LVEF justifies the increased level of SCD and heart transplant in males. Analysis of human induced-pluripotent stem cell (hiPSC)-derived cardiomyocytes (hiPSC-CMs) from DCM patients carrying rare variants in RBM20 have shown that pathogenic alterations in this gene may disorganize the sarcomeric complex [8][25]. The RBM20 hiPSC-CMs were defective in calcium handling machinery with prolonged levels in the cytoplasm and higher spike amplitude [8][25]. Indeed, this fact supports the malignant arrhythmogenic nature of the rare alterations in this gene [10], thereby requiring patients carrying these variants to be more closely followed together with the adoption of personalized preventive measures.
The stratification of SCD risk has been precluded by the use of heterogeneous subsets of patients with idiopathic DCM and by the use of risk models in which predictions are based on static parameters disregarding the disease course. Furthermore, it is important to consider that a large proportion of the RBM20-DCM patients published in the literature come from the same families, representing phenotypes of specific rare pathogenic alterations and possibly with an additional common genetic background that may add information to the aggressiveness of the disease. Similarly to other cardiomyopathies, disease expression may vary depending on site-specific alterations in RBM20, so accounting that information in future projects will bring clinical value to such work. Of note is that the aggressiveness of the disease has been demonstrated differently according to biological sex, which suggests that a different clinical approach should be applied accordingly. The current approach to personalized risk stratification for DCM is shifting towards a better characterization of the underlying etiology of DCM, in which genetic study has a paramount value. Family screening is mandatory among patients in order to identify asymptomatic DCM affected individuals at risk of SCD, which can be the first symptom of the disease.
References
- Zahr, H.C.; Jaalouk, D.E. Exploring the Crosstalk Between LMNA and Splicing Machinery Gene Mutations in Dilated Cardiomyopathy. Front. Genet. 2018, 9, 231.
- Murayama, R.; Kimura-Asami, M.; Togo-Ohno, M.; Yamasaki-Kato, Y.; Naruse, T.K.; Yamamoto, T.; Hayashi, T.; Ai, T.; Spoonamore, K.G.; Kovacs, R.J.; et al. Phosphorylation of the RSRSP stretch is critical for splicing regulation by RNA-Binding Motif Protein 20 (RBM20) through nuclear localization. Sci. Rep. 2018, 8, 8970.
- Filippello, A.; Lorenzi, P.; Bergamo, E.; Romanelli, M.G. Identification of nuclear retention domains in the RBM20 protein. FEBS Lett. 2013, 587, 2989–2995.
- Zerbino, D.R.; Achuthan, P.; Akanni, W.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Giron, C.G.; et al. Ensembl 2018. Nucleic Acids Res. 2018, 46, D754–D761.
- Lennermann, D.; Backs, J.; van den Hoogenhof, M.M.G. New Insights in RBM20 Cardiomyopathy. Curr. Heart Fail. Rep. 2020, 17, 234–246.
- Fochi, S.; Lorenzi, P.; Galasso, M.; Stefani, C.; Trabetti, E.; Zipeto, D.; Romanelli, M.G. The Emerging Role of the RBM20 and PTBP1 Ribonucleoproteins in Heart Development and Cardiovascular Diseases. Genes 2020, 11, 402.
- Guo, W.; Schafer, S.; Greaser, M.L.; Radke, M.H.; Liss, M.; Govindarajan, T.; Maatz, H.; Schulz, H.; Li, S.; Parrish, A.M.; et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat. Med. 2012, 18, 766–773.
- Wyles, S.P.; Li, X.; Hrstka, S.C.; Reyes, S.; Oommen, S.; Beraldi, R.; Edwards, J.; Terzic, A.; Olson, T.M.; Nelson, T.J. Modeling structural and functional deficiencies of RBM20 familial dilated cardiomyopathy using human induced pluripotent stem cells. Hum. Mol. Genet. 2016, 25, 254–265.
- Wilsbacher, L.D. Clinical Implications of the Genetic Architecture of Dilated Cardiomyopathy. Curr. Cardiol. Rep. 2020, 22, 170.
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424.
- Den Dunnen, J.T. Sequence Variant Descriptions: HGVS Nomenclature and Mutalyzer. Curr Protoc Hum. Genet. 2016, 90, 7–13.
- Den Dunnen, J.T.; Dalgleish, R.; Maglott, D.R.; Hart, R.K.; Greenblatt, M.S.; McGowan-Jordan, J.; Roux, A.F.; Smith, T.; Antonarakis, S.E.; Taschner, P.E. HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum. Mutat. 2016, 37, 564–569.
- Kobayashi, Y.; Yang, S.; Nykamp, K.; Garcia, J.; Lincoln, S.E.; Topper, S.E. Pathogenic variant burden in the ExAC database: An empirical approach to evaluating population data for clinical variant interpretation. Genome Med. 2017, 9, 13.
- Muller, R.D.; McDonald, T.; Pope, K.; Cragun, D. Evaluation of Clinical Practices Related to Variants of Uncertain Significance Results in Inherited Cardiac Arrhythmia and Inherited Cardiomyopathy Genes. Circ. Genom. Precis. Med. 2020, 13, e002789.
- Parikh, V.N.; Caleshu, C.; Reuter, C.; Lazzeroni, L.C.; Ingles, J.; Garcia, J.; McCaleb, K.; Adesiyun, T.; Sedaghat-Hamedani, F.; Kumar, S.; et al. Regional Variation in RBM20 Causes a Highly Penetrant Arrhythmogenic Cardiomyopathy. Circ. Heart Fail. 2019, 12, e005371.
- Briganti, F.; Sun, H.; Wei, W.; Wu, J.; Zhu, C.; Liss, M.; Karakikes, I.; Rego, S.; Cipriano, A.; Snyder, M.; et al. iPSC Modeling of RBM20-Deficient DCM Identifies Upregulation of RBM20 as a Therapeutic Strategy. Cell Rep. 2020, 32, 108117.
- Brauch, K.M.; Karst, M.L.; Herron, K.J.; de Andrade, M.; Pellikka, P.A.; Rodeheffer, R.J.; Michels, V.V.; Olson, T.M. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J. Am. Coll Cardiol 2009, 54, 930–941.
- Li, D.; Morales, A.; Gonzalez-Quintana, J.; Norton, N.; Siegfried, J.D.; Hofmeyer, M.; Hershberger, R.E. Identification of novel mutations in RBM20 in patients with dilated cardiomyopathy. Clin. Transl. Sci. 2010, 3, 90–97.
- Wells, Q.S.; Becker, J.R.; Su, Y.R.; Mosley, J.D.; Weeke, P.; D’Aoust, L.; Ausborn, N.L.; Ramirez, A.H.; Pfotenhauer, J.P.; Naftilan, A.J.; et al. Whole exome sequencing identifies a causal RBM20 mutation in a large pedigree with familial dilated cardiomyopathy. Circ. Cardiovasc Genet. 2013, 6, 317–326.
- Chami, N.; Tadros, R.; Lemarbre, F.; Lo, K.S.; Beaudoin, M.; Robb, L.; Labuda, D.; Tardif, J.C.; Racine, N.; Talajic, M.; et al. Nonsense mutations in BAG3 are associated with early-onset dilated cardiomyopathy in French Canadians. Can. J. Cardiol. 2014, 30, 1655–1661.
- Millat, G.; Bouvagnet, P.; Chevalier, P.; Sebbag, L.; Dulac, A.; Dauphin, C.; Jouk, P.S.; Delrue, M.A.; Thambo, J.B.; Le Metayer, P.; et al. Clinical and mutational spectrum in a cohort of 105 unrelated patients with dilated cardiomyopathy. Eur. J. Med Genet. 2011, 54, e570–e575.
- Refaat, M.M.; Lubitz, S.A.; Makino, S.; Islam, Z.; Frangiskakis, J.M.; Mehdi, H.; Gutmann, R.; Zhang, M.L.; Bloom, H.L.; MacRae, C.A.; et al. Genetic variation in the alternative splicing regulator RBM20 is associated with dilated cardiomyopathy. Heart Rhythm 2012, 9, 390–396.
- Gaertner, A.; Klauke, B.; Brodehl, A.; Milting, H. RBM20 mutations in left ventricular non-compaction cardiomyopathy. Pediatr Investig. 2020, 4, 61–63.
- Gaertner, A.; Klauke, B.; Felski, E.; Kassner, A.; Brodehl, A.; Gerdes, D.; Stanasiuk, C.; Ebbinghaus, H.; Schulz, U.; Dubowy, K.O.; et al. Cardiomyopathy-associated mutations in the RS domain affect nuclear localization of RBM20. Hum. Mutat 2020, 41, 1931–1943.
- Streckfuss-Bomeke, K.; Tiburcy, M.; Fomin, A.; Luo, X.; Li, W.; Fischer, C.; Ozcelik, C.; Perrot, A.; Sossalla, S.; Haas, J.; et al. Severe DCM phenotype of patient harboring RBM20 mutation S635A can be modeled by patient-specific induced pluripotent stem cell-derived cardiomyocytes. J. Mol. Cell Cardiol. 2017, 113, 9–21.
- Rebs, S.; Sedaghat-Hamedani, F.; Kayvanpour, E.; Meder, B.; Streckfuss-Bomeke, K. Generation of pluripotent stem cell lines and CRISPR/Cas9 modified isogenic controls from a patient with dilated cardiomyopathy harboring a RBM20 p.R634W mutation. Stem Cell Res. 2020, 47, 101901.
- Norton, N.; Siegfried, J.D.; Li, D.; Hershberger, R.E. Assessment of LMNA copy number variation in 58 probands with dilated cardiomyopathy. Clin. Transl. Sci. 2011, 4, 351–352.
- Norton, N.; Li, D.; Rieder, M.J.; Siegfried, J.D.; Rampersaud, E.; Zuchner, S.; Mangos, S.; Gonzalez-Quintana, J.; Wang, L.; McGee, S.; et al. Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am. J. Hum. Genet. 2011, 88, 273–282.
- Mates, J.; Mademont-Soler, I.; Del Olmo, B.; Ferrer-Costa, C.; Coll, M.; Perez-Serra, A.; Pico, F.; Allegue, C.; Fernandez-Falgueras, A.; Alvarez, P.; et al. Role of copy number variants in sudden cardiac death and related diseases: Genetic analysis and translation into clinical practice. Eur. J. Hum. Genet. 2018, 26, 1014–1025.
- Mates, J.; Mademont-Soler, I.; Fernandez-Falgueras, A.; Sarquella-Brugada, G.; Cesar, S.; Arbelo, E.; Garcia-Alvarez, A.; Jorda, P.; Toro, R.; Coll, M.; et al. Sudden Cardiac Death and Copy Number Variants: What Do We Know after 10 Years of Genetic Analysis? Forensic Sci. Int. Genet. 2020, 47, 102281.
- Haas, J.; Frese, K.S.; Peil, B.; Kloos, W.; Keller, A.; Nietsch, R.; Feng, Z.; Muller, S.; Kayvanpour, E.; Vogel, B.; et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J. 2015, 36, 1123–1135.
- Kayvanpour, E.; Sedaghat-Hamedani, F.; Amr, A.; Lai, A.; Haas, J.; Holzer, D.B.; Frese, K.S.; Keller, A.; Jensen, K.; Katus, H.A.; et al. Genotype-phenotype associations in dilated cardiomyopathy: Meta-analysis on more than 8000 individuals. Clin. Res. Cardiol. 2017, 106, 127–139.
- Hey, T.M.; Rasmussen, T.B.; Madsen, T.; Aagaard, M.M.; Harbo, M.; Molgaard, H.; Moller, J.E.; Eiskjaer, H.; Mogensen, J. Pathogenic RBM20-Variants Are Associated With a Severe Disease Expression in Male Patients With Dilated Cardiomyopathy. Circ. Heart Fail. 2019, 12, e005700.
- Van den Hoogenhof, M.M.G.; Beqqali, A.; Amin, A.S.; van der Made, I.; Aufiero, S.; Khan, M.A.F.; Schumacher, C.A.; Jansweijer, J.A.; van Spaendonck-Zwarts, K.Y.; Remme, C.A.; et al. RBM20 Mutations Induce an Arrhythmogenic Dilated Cardiomyopathy Related to Disturbed Calcium Handling. Circulation 2018, 138, 1330–1342.
- Nielsen, J.B.; Fritsche, L.G.; Zhou, W.; Teslovich, T.M.; Holmen, O.L.; Gustafsson, S.; Gabrielsen, M.E.; Schmidt, E.M.; Beaumont, R.; Wolford, B.N.; et al. Genome-wide Study of Atrial Fibrillation Identifies Seven Risk Loci and Highlights Biological Pathways and Regulatory Elements Involved in Cardiac Development. Am. J. Hum. Genet. 2018, 102, 103–115.
- Roselli, C.; Chaffin, M.D.; Weng, L.C.; Aeschbacher, S.; Ahlberg, G.; Albert, C.M.; Almgren, P.; Alonso, A.; Anderson, C.D.; Aragam, K.G.; et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat. Genet. 2018, 50, 1225–1233.





