
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | John Bladen | + 2332 word(s) | 2332 | 2021-03-09 06:45:04 | | | |
| 2 | Bruce Ren | Meta information modification | 2332 | 2021-03-16 02:29:30 | | | | |
| 3 | Bruce Ren | Meta information modification | 2332 | 2021-03-16 02:30:46 | | |
Video Upload Options
Blepharophimosis, ptosis, and epicanthus inversus syndrome (BPES) is a craniofacial disorder caused by heterozygous variants of the forkhead box L2 (FOXL2) gene. It shows autosomal dominant inheritance but can also occur sporadically. Depending on the mutation, two phenotypic subtypes have been described, both involving the same craniofacial features: type I, which is associated with premature ovarian failure (POF), and type II, which has no systemic features.
1. Introduction
Blepharophimosis, ptosis, and epicanthus inversus syndrome (BPES; OMIM #110100) is a rare autosomal dominant disease, with an estimated prevalence of 1 in 50,000 births, primarily affecting the development of the mid-face structures [1]. The four major clinical signs are dysplasia of the eyelids with shortening of the horizontal fissures (blepharophimosis), droopy upper lids reducing the vertical palpebral aperture (ptosis), bilateral skin fold arising from the medial lower eyelid ascending to the upper lid (epicanthus inversus), and an increased distance between the medial canthi (telecanthus). Two main phenotypes of BPES have emerged, and each harbour the four key ocular signs: (i) type I (BPES-I), which is also associated with premature ovarian failure (POF) involving secondary amenorrhoea before 40 years of age, leading to early menopause and infertility, and (ii) type II (BPES-II) with no systemic associations.
BPES can be caused by heterozygous variants involving the forkhead box L2 (FOXL2) gene, which encodes for a transcription factor expressed predominantly in the developing mesenchyme of eyelids and ovaries [2]. In mice, Foxl2 expression is localised to the protruding ridges of the developing eyelids and in ovarian follicular cells. Up to 75% of affected individuals may have detectable FOXL2 mutation, leading to haploinsufficiency [3][4][5][6]. Transmission of BPES-I is usually by affected males as fertility in affected females is reduced due to ovarian dysfunction. BPES-II can have transmission occurring through both males and females.
2. Genetics of BPES
2.1. FOXL2 Gene
FOXL2 is a single-exon gene consisting of 2.9 kb (NM_023067.4) located on chromosome 3q22.3. The transcribed protein is 376 amino acids and belongs to the family of forkhead/winged helix transcription factors. FOXL2 regulates a number of genes that control cellular processes including inflammation, transcription, proteolysis, apoptosis, and steroidogenesis including gonadotrophins [7][8]. It is highly conserved among species, with 100% of homology for human, mouse, rat, cow, goat, pig, and rabbit, and consists of a 110 amino acid forkhead DNA-binding domain at position 54 to 148 [9]. It also contains a strictly conserved poly-alanine tract of 14 amino acids between position 221 and 234, whose role remains unknown; however, it is a hotspot for expansions from 14 to 24 alanine residues accounting for ≈30% of all intragenic FOXL2 pathogenic variants leading to predominantly BPES-II [6][9].
Haploinsufficiency of FOXL2 remains the only reported cause of BPES, and the first autosomal gene implicated in syndromic POF [2]. FOXL2 can be disrupted by intragenic mutations as well as larger genomic deletions involving the gene locus [10]. More than 250 variants are associated with BPES [11]: intragenic mutations of FOXL2 account for 81% and can be subdivided into indel frameshift (44%), in-frame deletions (33%), nonsense (12%), missense (11%), and duplications [11]. Whole-gene deletions and larger sub-microscopic deletions encompassing FOXL2 and neighbouring genes represent 12% and 5% of molecularly confirmed cases, respectively [11].
At least 460 patients have been reported with FOXL2-related BPES [1][6][10][12][13][14][15][16][17][18][19][20][21][22][23][24][25][26][27][28][29][30][31][32][33][34][35][36][37][38][39][40][41][42][43][44][45][46][47][48][49][50][51][52][53][54][55][56][57][58][59][60][61][62]. The most common variants affect two intragenic regions (Figure 1): (i) the poly-alanine region with c.663_692dup p.(Ala221_Ala231dup) reported in 12 patients (four BPES-II cases and eight with undefined type) [6][14][16][30][31][32], c.664_693dup p.(Ala222_Ala231dup) reported in five patients (two BPES-II, two BPES-I, and one with undefined type), and c.672_701dup p.(Ala225_Ala234dup), which was reported in at least 80 patients (24 BPES-II, 2 BPES-I, and 54 with undefined type) [6][14][16][22][26][30][31][32][54][61][63], and (ii) the poly-proline region, which encompasses amino acids at positions 284 to 292, has two duplications variants: c.843_859dup p.(Pro287Argfs*75) reported in 46 patients (3 BPES-I, 2 BPES-II, and 41 with undefined type) [6][14][16][31][32][63] and c.855_871dup p.(His291Argfs*71) in 15 patients (3 BPES-I and 12 undefined type) [6][14][16][24][38][63], and the deletion c.855_871del p.(Pro287Alafs*71) in 5 patients (3 BPES-I, 1 BPES-II, and 1 with undefined type) [6][14][34][40][63].
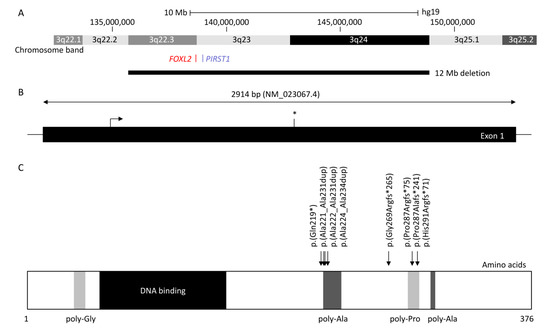
A classification (groups A-H) of intragenic variants in ≈500 FOXL2 cases was previously proposed by De Baere et al. in order to determine a genotype–phenotype correlation [6]. However, as ovarian function was largely unavailable due to the young age or sex of the patient, this was not possible. A tendency was observed with variants leading to an expanded poly-alanine region, such as c.663_692dup p.(Ala221_Ala231dup), c.664_693dup p.(Ala222_Ala231dup), and c.672_701dup p.(Ala225_Ala234dup), which are mostly associated with BPES-II [6][14][16][22][26][30][31][32][43][45][51][54][61][63]. Interfamilial variability was observed for these variants, where some cases are reported to be associated with BPES-I. Of note, one family reported a BPES-II-affected mother and her BPES-I-affected daughter, both carrying c.822C>G p.(Tyr274*), revealing possible intrafamilial phenotypic variability [6].
2.2. Intragenic Variants in FOXL2
As FOXL2 is composed of one exon, it may be resistant to nonsense mediated decay (NMD) as in other cases [64]. Thus, null variants in FOXL2 will result in a truncated protein leading to partial or complete loss of the forkhead domain and poly-alanine tract [6] or in a shorter protein without the N-terminal region, due to a re-initiation of translation, as observed in COS-7 cell lines transfected with the c.157C>T p.(Gln53*) construct [65]. Whereas duplications within or downstream of the forkhead domain can be predicted to result in an extended protein, mutations involving only part of the forkhead domain may give rise to haploinsufficiency and BPES-II by reducing the transactivation activity of the gene without affecting its DNA binding [66]. The effect of missense mutations can vary depending on their location, which may be in a functionally important region as the gene is highly conserved [66]. Most are mapped to the forkhead DNA binding domain, and these are likely to be pathogenic. However, as missense mutations have been reported in both BPES-I and -II, no prediction can be made regarding the genotype–phenotype correlation [6].
2.3. Poly-Alanine Tract Expansion Variants
Studies have confirmed the existence of a mutational hotspot in the poly-alanine tract of FOXL2 in families of different ethnicities [6][67]. This highly conserved region consists of 14 Alanine residues, and the secondary protein structure is predicted to be an α helix, which may become distorted when mutated and disrupt an essential function [6]. Poly-alanine tract expansion is the most common mutation to have been described in BPES-II [67]. Eight different alanine tract expansions (c.663_692dup30 p.(Ala221_Ala231dup), c.664_693dup30 p.(Ala222_Ala231dup), c.664_701dup p.(Ala222_Ala234dup), c.667_702dup p.(Ala223_Ala234), c.672_701dup30 p.(Ala225_Ala234dup), c.684_698dup p.(Ala228_Ala232dup), c.684_698trip15 p.(Ala228_Ala232trip), c.696_728dup p.(Ala232_Ala243dup)) have been described, with the most common consisting of a repeat of 30 bases [6][14][16][22][24][30][31][32][41][54][55][61][63]. These expansions may be caused by slippage of DNA polymerase when duplicating trinucleotide repeats, accounting for about 33% of all intragenic mutations in BPES overall [6][11][41][67]. Whilst expansions are more likely to be associated with BPES-II (without ovarian involvement) and truncated proteins are correlated with BPES-I (with ovarian involvement) [68], some poly-alanine expansions such as c.664_693dup30 p.(Ala222_Ala231dup) and c.672_701dup p.(Ala224_Ala234dup) have resulted in some degree of ovarian dysfunction [14][45]. There has only been one autosomal recessive consanguineous Indian family with evidence of a homozygous poly-alanine tract expansion, c.684_698dup p.(Ala228_Ala232dup), with ovarian failure; segregated carrier parents and siblings were not affected. As such, BPES is mainly regarded as an autosomal dominant disorder [41].
Further functional analyses have been performed to study mutation consequences at a cellular level. Luciferase assays revealed some nonsense variants, such as p.(Glu19*), produced a shorter protein with an alternative initiation codon and formed nuclear aggregates, while wild-type protein was diffuse in the nucleus [65]. Caburet et al. showed protein mislocalisation from the nucleus to the cytoplasm with mutated FOXL2 poly-alanine tract expansions leading to its cytoplasmic aggregation [69]. Moreover, these poly-alanine tract expansions lead to decreased expression of several genes involved in apoptosis, transcriptional regulation, mediation of inflammation, cholesterol metabolism, and reactive oxygen species detoxification [70]. Two models were suggested to explain BPES phenotype with or without POF: (1) a higher dose of functional FOXL2 might be required to target promoters in the developing eyelid than in ovarian follicular cells, or (2) the number of FOXL2 binding sites in promoters is the same in both tissues, but aggregation and mislocalisation of mutant protein are stronger in the eyelids than ovaries, due to different tissue-specific proteomics [70]. Some missense variants, such as c.931C>T p.(His311Tyr) associated with BPES-I [6] or with undefined type due to the young age of female patients [62], were also reported to affect the expression of targeted genes, such as the steroidogenic acute regulatory gene (STAR, OMIM 600617) [62].
2.4. Chromosomal Translocations and Involvement of FOXL2 Regulatory Genes
Translocation breakpoints in chromosome 3 close to the FOXL2 gene and FOXL2 regulatory genes such as PISRT1 in nine patients carrying t(1;3)(p21;q22), t(1;3) associated with a 1.2 Mb deletion in 3q23 upstream of the FOXL2 transcription unit, t(2;3)(q33;q23), t(3;4)(q23;p15), t(3;7)(q23;q32), t(3;11)(q22.3;q14.1), t(3;15)(q23;q25), t(3;20)(q22;q13), and t(3;21)(q23;q22.1) were shown to be correlated with BPES [6][12][25][48][52][59][71][72]. Only the patient with t(3;11)(q22.3;q14.1) was known to have BPES-I [48]. Although the FOXL2 gene itself carried no mutation, positional effects are prevalent in human genetic diseases involving transcriptional factors, for example, PAX6 and PITX2 genes, which are involved in aniridia and Axenfeld–Rieger syndrome, respectively [73].
Genomic alterations in loci outside of the FOXL2 region such as deletion of upstream or downstream regulatory regions close to FOXL2 or PISTR1 deletion have also been found to account for about 5% of BPES [53]. Larger deletions encompassing 3q22.3-3q24 are associated with an undefined type of BPES, Dandy–Walker malformation, and Wisconsin syndrome [47], whereas smaller deletions of FOXL2 or PISTR1 gene lead to just undefined BPES [63]. Upstream regions of FOXL2 and PISTR1 genes are highly conserved in goat, mouse, and human [74], and their deletion leads to polled goats due to PIS (polled intersex syndrome) mutation [75]. This model is characterised by cranio-facial defects, female sterility, and XX sex reversal, associated with decreased expression level of FOXL2 and PISTR1 in the ovaries [75]. Luciferase assay revealed the identified genomic deletions in loci outside of the FOXL2 region affect gene expression in ovarian cell lines [21]. Rearrangements, such as large chromosomal deletion or translocation occurring in these regions, can dissociate the transcription unit from its regulatory elements, resulting in the same phenotype as intragenic mutations [5]. Total and partial gene deletions as well as microdeletions mapping upstream and downstream of FOXL2 have been found in cases of sporadic and familial cases of BPES [5]. These deletion points are scattered and lie in transcription factor-binding sites and the goat PIS locus requiring further investigation to fully understand the intergenic regulatory elements [5]. Deletions were shown to be conserved between different generations of affected family members, revealing meiotic stability. A number of microdeletions, such as a 197 kb deletion upstream of FOXL2, have been correlated with BPES-like disorders associated with microcephaly and intellectual disability [13].
2.5. FOXL2 and Primary Ovarian Failure (POF)
FOXL2 is the earliest known but not the sole regulator of sex differentiation in mammals [76]. It is involved in foetal development as well as maintenance of the mature ovary. In the postnatal ovary, FOXL2 supports follicular growth. Ablation of FOXL2 in mice led to atresia of the oocytes with no maturation of secondary follicles. It has been shown that the STAR protein, which is a marker of granulosa cell differentiation, is a direct target of FOXL2, acting as a repressor of STAR. It was concluded that the entire alanine/proline-rich carboxyl terminus is important for the repressor activity of FOXL2 and that truncating variants may preferentially lead to BPES and ovarian dysfunction by accelerated differentiation of granulosa cells and secondary depletion of the primordial follicle pool. The identification of a considerable number of ovarian FOXL2 targets may be essential to reveal more insights into phenotypic effects of FOXL2 pathogenic variants in the adult ovary. Digenic inheritance might contribute to POF associated with BPES through a synergistic effect of FOXL2 mutations and other genes involved in ovarian function. This may also explain the apparent pleiotropism of FOXL2 mutations.
The phenomena of waxing and waning gonadotrophin levels which suggest infertility may be partially reversible in BPES type I. Varying gonadotrophin levels in female patients with BPES type I may not necessarily meet the diagnostic criteria for POF. POF is commonly defined as the presence of four or more months of secondary amenorrhea, postmenopausal levels of follicle-stimulating hormone (FSH; >40 IU/L) all before the age of 40 years [77]. However, there is no universal definition of POF, and coupled with the large normal variation in ovarian reserve, it can be difficult to make a diagnosis of a POF [78][79]. Moreover, spontaneous pregnancies and pregnancies post-stimulation with gonadotrophins have been reported in individuals with FOXL2 mutations and in women with POF alone [22][80].
References
- Chawla, B.; Bhadange, Y.; Dada, R.; Kumar, M.; Sharma, S.; Bajaj, M.S.; Pushker, N.; Chandra, M.; Ghose, S. Clinical, Radiologic, and Genetic Features in Blepharophimosis, Ptosis, and Epicanthus Inversus Syndrome in the Indian Population. Investig. Opthalmol. Vis. Sci. 2013, 54, 2985–2991.
- Cocquet, J.; Pailhoux, E.; Jaubert, F.; Servel, N.; Xia, X.; Pannetier, M.; De Baere, E.; Messiaen, L.; Cotinot, C.; Fellous, M.; et al. Evolution and Expression of FOXL2. J. Med. Genet. 2002, 39, 916–921.
- Chen, H. Blepharophimosis, Ptosis, and Epicanthus Inversus Syndrome. In Atlas of Genetic Diagnosis and Counseling; Springer: New York, NY, USA, 2012; pp. 233–238.
- Fokstuen, S.; Antonarakis, S.E.; Blouin, J.-L. FOXL2-Mutations in Blepharophimosis-Ptosis-Epicanthus Inversus Syndrome (BPES); Challenges for Genetic Counseling in Female Patients. Am. J. Med. Genet. Part A 2002, 117, 143–146.
- Beysen, D.; Raes, J.; Leroy, B.P.; Lucassen, A.; Yates, J.R.W.; Clayton-Smith, J.; Ilyina, H.; Brooks, S.S.; Christin-Maitre, S.; Fellous, M.; et al. Deletions Involving Long-Range Conserved Nongenic Sequences Upstream and Downstream of FOXL2 as a Novel Disease-Causing Mechanism in Blepharophimosis Syndrome. Am. J. Hum. Genet. 2005, 77, 205–218.
- De Baere, E.; Beysen, D.; Oley, C.; Lorenz, B.; Cocquet, J.; De Sutter, P.; Devriendt, K.; Dixon, M.; Fellous, M.; Fryns, J.-P.; et al. FOXL2 and BPES: Mutational Hotspots, Phenotypic Variability, and Revision of the Genotype-Phenotype Correlation. Am. J. Hum. Genet. 2003, 72, 478–487.
- Batista, F.; Vaiman, D.; Dausset, J.; Fellous, M.; Veitia, R.A. Potential Targets of FOXL2, a Transcription Factor Involved in Craniofacial and Follicular Development, Identified by Transcriptomics. Proc. Natl. Acad. Sci. USA 2007, 104, 3330–3335.
- Leung, D.T.; Fuller, P.J.; Chu, S. Impact of FOXL2 Mutations on Signaling in Ovarian Granulosa Cell Tumors. Int. J. Biochem. Cell Biol. 2016, 72, 51–54.
- Cocquet, J.; De Baere, E.; Gareil, M.; Pannetier, M.; Xia, X.; Fellous, M.; Veitia, R. Structure, Evolution and Expression of the FOXL2 Transcription Unit. Cytogenet. Genome Res. 2003, 101, 206–211.
- D’Haene, B.; Nevado, J.; Pugeat, M.; Pierquin, G.; Lowry, R.; Reardon, W.; Delicado, A.; García-Miñaur, S.; Palomares, M.; Courtens, W.; et al. FOXL2 Copy Number Changes in the Molecular Pathogenesis of BPES: Unique Cohort of 17 Deletions. Hum. Mutat. 2010, 31, E1332–E1347.
- Beysen, D.; De Paepe, A.; De Baere, E. FOXL2 Mutations and Genomic Rearrangements in BPES. Hum. Mutat. 2008, 30, 158–169.
- Alao, M.; Laleye, A.; Lalya, F.; Hans, C.; Abramovicz, M.; Morice-Picard, F.; Arveiler, B.; Lacombe, D.; Rooryck, C. Blepharophimosis, Ptosis, Epicanthus Inversus Syndrome with Translocation and Deletion at Chromosome 3q23 in a Black African female. Eur. J. Med. Genet. 2012, 55, 630–634.
- Bertini, V.; Valetto, A.; Baldinotti, F.; Azzarà, A.; Cambi, F.; Toschi, B.; Giacomina, A.; Gatti, G.L.; Gana, S.; Caligo, M.A.; et al. Blepharophimosis, Ptosis, Epicanthus Inversus Syndrome: New Report with a 197-kb Deletion Upstream of FOXL2 and Review of the Literature. Mol. Syndr. 2019, 10, 147–153.
- Beysen, D.; De Jaegere, S.; Amor, D.; Bouchard, P.; Christin-Maitre, S.; Fellous, M.; Touraine, P.; Grix, A.W.; Hennekam, R.; Meire, F.; et al. Identification of 34 Novel and 56 Known FOXL2 Mutations in Patients with Blepharophimosis Syndrome. Hum. Mutat. 2008, 29, E205–E219.
- Bouman, A.; Van Haelst, M.; Van Spaendonk, R. Blepharophimosis–Ptosis–Epicanthus Inversus Syndrome Caused by a 54-KB Microdeletion in a FOXL2 Cis-Regulatory Element. Clin. Dysmorphol. 2018, 27, 58–62.
- Chacon-Camacho, O.F.; Salgado-Medina, A.; Alcaraz-Lares, N.; López-Moreno, D.; Barragan-Arevalo, T.; Nava-Castañeda, A.; Rodríguez-Uribe, G.; Lieberman, E.; Rodríguez-Cabrera, L.; Angel, A.G.-D.; et al. Clinical Characterization and Identification of Five Novel FOXL2 Pathogenic Variants in a Cohort of 12 Mexican Subjects with the Syndrome of Blepharophimosis-Ptosis-Epicanthus Inversus. Gene 2019, 706, 62–68.
- Chai, P.; Li, F.; Fan, J.; Jia, R.; Zhang, H.; Fan, X. Functional Analysis of a Novel FOXL2 Indel Mutation in Chinese Families with Blepharophimosis-Ptosis-Epicanthus Inversus Syndrome Type, I. Int. J. Biol. Sci. 2017, 13, 1019–1028.
- Chouchene, I.; Derouiche, K.; Chaabouni, A.; Cherif, L.; Amouri, A.; Largueche, L.; Abdelhak, S.; El Matri, L. Identification of a Novel Mutation inFOXL2Gene That Leads to Blepharophimosis Ptosis Epicanthus Inversus and Telecanthus Syndrome in a Tunisian Consanguineous Family. Genet. Test. Mol. Biomarkers 2010, 14, 145–148.
- Corrêa, F.J.S.; Tavares, A.B.; Pereira, R.W.; Abrão, M.S. A New FOXL2 Gene Mutation in a Woman with Premature Ovarian Failure and Sporadic Blepharophimosis-Ptosis-Epicanthus Inversus Syndrome. Fertil. Steril. 2010, 93, 1006.e3–1006.e6.
- Dean, S.J.; Holden, K.R.; Dwivedi, A.; Dupont, B.R.; Lyons, M.J. Acquired Microcephaly in Blepharophimosis-Ptosis-Epicanthus Inversus Syndrome Because of an Interstitial 3q22.3q23 Deletion. Pediatr. Neurol. 2014, 50, 636–639.
- D’Haene, B.; Attanasio, C.; Beysen, D.; Dostie, J.; Lemire, E.; Bouchard, P.; Field, M.; Jones, K.; Lorenz, B.; Menten, B.; et al. Disease-Causing 7.4 kb Cis-Regulatory Deletion Disrupting Conserved Non-Coding Sequences and Their Interaction with the FOXL2 Promotor: Implications for Mutation Screening. PLoS Genet. 2009, 5, e1000522.
- Fan, J.; Zhou, Y.; Huang, X.; Zhang, L.; Yao, Y.; Song, X.; Chen, J.; Hu, J.; Ge, S.; Song, H.; et al. The Combination of Polyalanine Expansion Mutation and a Novel Missense Substitution in Transcription Factor FOXL2 Leads to Different Ovarian Phenotypes in Blepharophimosis-Ptosis-Epicanthus Inversus Syndrome (BPES) Patients. Hum. Reprod. 2012, 27, 3347–3357.
- Fan, J.-Y.; Han, B.; Qiao, J.; Liu, B.-L.; Ji, Y.-R.; Ge, S.-F.; Song, H.-D.; Fan, X.-Q. Functional Study on a Novel Missense Mutation of the Transcription Factor FOXL2 Causes Blepharophimosis-Ptosis-Epicanthus Inversus Syndrome (BPES). Mutagenesis 2010, 26, 283–289.
- Fan, J.-Y.; Wang, Y.-F.; Han, B.; Ji, Y.-R.; Song, H.-D.; Fan, X.-Q. FOXL2 Mutations in Chinese Families with Blepharophimosis Syndrome (BPES). Transl. Res. 2011, 157, 48–52.
- González-González, C.; Garcia-Hoyos, M.; Calzón, R.H.; Díaz, C.A.; Fanego, C.G.; Sánchez, I.L.; Sánchez-Escribano, F. Microdeletion Found by Array-CGH in Girl with Blepharophimosis Syndrome and Apparently Balanced Translocation t(3;15) (q23;q25). Ophthalmic Genet. 2011, 33, 107–110.
- Gulati, R.; Verdin, H.; Halanaik, D.; Bhat, B.V.; De Baere, E. Co-Occurrence of Congenital Hydronephrosis and FOXL2-Associated Blepharophimosis, Ptosis, Epicanthus Inversus Syndrome (BPES). Eur. J. Med. Genet. 2014, 57, 576–578.
- Haghighi, A.; Verdin, H.; Haghighi-Kakhki, H.; Piri, N.; Gohari, N.S.; De Baere, E. Missense Mutation outside the Forkhead Domain of FOXL2 Causes a Severe Form of BPES Type II. Mol. Vis. 2012, 18, 211–218.
- Hu, J.; Ke, H.; Luo, W.; Yang, Y.; Liu, H.; Li, G.; Qin, Y.; Ma, J.; Zhao, S. A novel FOXL2 Mutation in Two Infertile Patients with Blepharophimosis–Ptosis–Epicanthus Inversus Syndrome. J. Assist. Reprod. Genet. 2020, 37, 223–229.
- Hu, S.; Guo, J.; Wang, B.; Wang, J.; Zhou, Z.; Zhou, G.; Ding, X.; Ma, X.; Qi, Y. Genetic Analysis of the FOXL2 Gene Using Quantitative Real-Time PCR in Chinese Patients with Blepharophimosis-Ptosis-Epicanthus In-versus Syndrome. Mol. Vis. 2011, 17, 436–442.
- Jiang, H.; Huang, X.; Su, Z.; Rao, L.; Wu, S.; Zhang, T.; Li, K.; Quan, Q.; Zhang, K. Genetic Analysis of the Fork-head Transcriptional Factor 2 Gene in Three Chinese Families with Blepharophimosis Syndrome. Mol. Vis. 2013, 19, 418–423.
- Kaur, I.; Hussain, A.; Naik, M.N.; Murthy, R.; Honavar, S.G. Mutation spectrum of Fork-Head Transcriptional Factor Gene (FOXL2) in Indian Blepharophimosis Ptosis Epicanthus Inversus Syndrome (BPES) patients. Br. J. Ophthalmol. 2011, 95, 881–886.
- Krepelova, A.; Simandlova, M.; Vlckova, M.; Kuthan, P.; Vincent, A.L.; Liskova, P. Analysis of FOXL2 Detects Three Novel Mutations and an Atypical Phenotype of Blepharophimosis-Ptosis-Epicanthus Inversus Syndrome. Clin. Exp. Ophthalmol. 2016, 44, 757–762.
- Kumar, A.; Babu, M.; Raghunath, A.; Venkatesh, C.P. Genetic Analysis of a Five Generation Indian Family with BPES: A Novel Missense Mutation (p.Y215C). Mol. Vis. 2004, 10, 445–449.
- Leon-Mateos, A.; Ginarte, M.; Ruiz-Ponte, C.; Carracedo, A.; Toribio, J. Blepharophimosis Ptosis Epicanthus Inversus Syndrome (BPES). Int. J. Dermatol. 2007, 46, 61–63.
- Li, F.; Chai, P.; Fan, J.; Wang, X.; Lu, W.; Li, J.; Ge, S.; Jia, R.; Zhang, H.; Fan, X. A Novel FOXL2 Mutation Implying Blepharophimosis-Ptosis-Epicanthus Inversus Syndrome Type, I. Cell. Physiol. Biochem. 2018, 45, 203–211.
- Li, H.; Gu, Y. Genetic and Functional Analyses of Two Missense Mutations in the Transcription Factor FOXL2 in Two Chinese Families with Blepharophimosis–Ptosis–Epicanthus Inversus Syndrome. Genet. Test. Mol. Biomarkers 2018, 22, 585–592.
- Lim, B.C.; Park, W.Y.; Seo, E.-J.; Kim, K.J.; Hwang, Y.S.; Chae, J.H. De Novo Interstitial Deletion of 3q22.3-q25.2 Encompassing FOXL2, ATR, ZIC1, and ZIC4 in a Patient with Blepharophimosis/Ptosis/Epicanthus Inversus Syndrome, Dandy-Walker Malformation, and Global Developmental Delay. J. Child Neurol. 2011, 26, 615–618.
- Lin, W.-D.; Chou, I.-C.; Lee, N.-C.; Wang, C.-H.; Hwu, W.-L.; Lin, S.-P.; Chao, M.-C.; Tsai, Y.; Tsai, F.-J. FOXL2 Mutations in Taiwanese Patients with Blepharophimosis, Ptosis, Epicanthus Inversus Syndrome. Clin. Chem. Lab. Med. 2010, 48, 485–488.
- Martínez-Aguayo, A.; Poggi, H.; Cattani, A.; Molina, M.; Romeo, E.; Lagos, M. A Novel Insertion in the FOXL2 Gene in a Chilean Patient with Blepharophimosis Ptosis Epicanthus Inversus Syndrome Type I. J. Pediatr. Endocrinol. Metab. 2014, 27, 181–184.
- Méduri, G.; Bachelot, A.; Duflos, C.; Bständig, B.; Poirot, C.; Genestie, C.; Veitia, R.; De Baere, E.; Touraine, P. FOXL2 Mutations Lead to Different Ovarian Phenotypes in BPES Patients: Case Report. Hum. Reprod. 2010, 25, 235–243.
- Nallathambi, J.; Moumné, L.; De Baere, E.; Beysen, D.; Usha, K.; Sundaresan, P.; Veitia, R.A. A Novel Polyalanine Expansion in FOXL2: The First Evidence for a Recessive Form of the Blepharophimosis Syndrome (BPES) Associated with Ovarian Dysfunction. Qual. Life Res. 2006, 121, 107–112.
- Ng, J.K.; Stout, A.U.; Aaby, A.A.; Ng, J.D. Blepharophimosis Syndrome with Absent Tear Production. Ophthalmic Plast. Reconstr. Surg. 2015, 31, e62.
- Ni, F.; Wen, Q.; Wang, B.; Zhou, S.; Wang, J.; Mu, Y.; Ma, X.; Cao, Y. Mutation Analysis of FOXL2 Gene in Chinese Patients with Premature Ovarian Failure. Gynecol. Endocrinol. 2009, 26, 246–249.
- Niu, B.-B.; Tang, N.; Xu, Q.; Chai, P.-W. Genomic Disruption of FOXL2 in Blepharophimosis-Ptosis-Epicanthus Inversus Syndrome Type 2. Chin. Med. J. 2018, 131, 2380–2383.
- Nuovo, S.; Passeri, M.; Di Benedetto, E.; Calanchini, M.; Meldolesi, I.; Di Giacomo, M.C.; Petruzzi, D.; Piemontese, M.R.; Zelante, L.; Sangiuolo, F.; et al. Characterization of Endocrine Features and Genotype–Phenotypes Correlations in Blepharophimosis–Ptosis–Epicanthus Inversus Syndrome Type 1. J. Endocrinol. Investig. 2015, 39, 227–233.
- Raile, K.; Stobbe, H.; Tröbs, R.B.; Kiess, W.; Pfäffle, R. A New Heterozygous Mutation of the FOXL2 Gene Is Associated with a Large Ovarian Cyst and Ovarian Dysfunction in an Adolescent Girl with Blepharophimosis/Ptosis/Epicanthus Inversus Syndrome. Eur. J. Endocrinol. 2005, 153, 353–358.
- Ramineni, A.; Coman, D. De Novo 3q22.3q24 Microdeletion in a Patient with Blepharophimosis–Ptosis–Epicanthus Inversus Syndrome, Dandy-Walker Malformation, and Wisconsin Syndrome. Child Neurol. Open 2016, 3, 2329048–16666362.
- Schlade-Bartusiak, K.; Brown, L.; Lomax, B.; Bruyère, H.; Gillan, T.; Hamilton, S.; McGillivray, B.; Eydoux, P. BPES With Atypical Premature Ovarian Insufficiency, and Evidence of Mitotic Recombination, in a Woman with Trisomy X and a Translocation t(3;11) (q22.3;q14.1). Am. J. Med. Genet. Part A 2012, 158A, 2322–2327.
- Settas, N.; Anapliotou, M.; Kanavakis, E.; Fryssira, H.; Sofocleous, C.; Dacou-Voutetakis, C.; Chrousos, G.P.; Voutetakis, A. A Novel FOXL2 Gene Mutation and BMP15 Variants in a Woman with Primary Ovarian Insufficiency and Blepharophimosis–Ptosis–Epicanthus Inversus Syndrome. Menopause 2015, 22, 1264–1268.
- Tan, H.; Yang, P.; Li, H.; Pan, Q.; Liang, D.; Wu, L. A Novel FOXL2 Mutation in a Chinese Family with Blepharophimosis, Ptosis, Epicanthus Inversus Syndrome. Hum. Genome Var. 2015, 2, 15008.
- Tang, S.; Wang, X.; Lin, L.; Sun, Y.; Wang, Y.; Yu, H. Mutation Analysis of the FOXL2 Gene in Chinese Patients with Blepharophimosis–Ptosis–Epicanthus Inversus Syndrome. Mutagenesis 2006, 21, 35–39.
- Tzschach, A.; Kelbova, C.; Weidensee, S.; Peters, H.; Ropers, H.-H.; Ullmann, R.; Erdogan, F.; Jurkatis, J.; Menzel, C.; Kalscheuer, V.; et al. Blepharophimosis-Ptosis-Epicanthus Inversus Syndrome in a Girl with Chromosome Translocation t(2;3) (q33;q23). Ophthalmic Genet. 2008, 29, 37–40.
- Verdin, H.; D’Haene, B.; Beysen, D.; Novikova, Y.; Menten, B.; Sante, T.; Lapunzina, P.; Nevado, J.; Carvalho, C.M.B.; Lupski, J.R.; et al. Microhomology-Mediated Mechanisms Underlie Non-Recurrent Disease-Causing Microdeletions of the FOXL2 Gene or Its Regulatory Domain. PLoS Genet. 2013, 9, e1003358.
- Wang, J.; Liu, J.; Zhang, Q. FOXL2 Mutations in Chinese Patients with Blepharophimosis-Ptosis-Epicanthus Inversus Syndrome. Mol. Vis. 2007, 13, 108–113.
- Xu, Y.; Lei, H.; Dong, H.; Zhang, L.; Qin, Q.; Gao, J.; Zou, Y.; Yan, X. FOXL2 Gene Mutations and Blepharophimosis-Ptosis-Epicanthus Inversus Syndrome (BPES): A Novel Mutation Detected in a Chinese Family and a Statistic Model for Summarizing Previous Reported Records. Mutagenesis 2009, 24, 447–453.
- Xue, M.; Zheng, J.; Zhou, Q.; Hejtmancik, J.F.; Wang, Y.; Li, S. Novel FOXL2 Mutations in Two Chinese Families with Blepharophimosis-Ptosis-Epicanthus Inversus Syndrome. BMC Med. Genet. 2015, 16, 73.
- Yang, L.; Li, T.; Xing, Y. Identification of a Novel FOXL2 Mutation in a Single Family with Both Types of Blepharophimosis-Ptosis-Epicanthus Inversus Syndrome. Mol. Med. Rep. 2017, 16, 5529–5532.
- Yang, X.-W.; He, W.-B.; Gong, F.; Li, W.; Li, X.-R.; Zhong, C.-G.; Lu, G.-X.; Lin, G.; Du, J.; Tan, Y.-Q. Novel FOXL2 Mutations Cause Blepharophimosis-Ptosis-Epicanthus Inversus Syndrome with Premature Ovarian Insufficiency. Mol. Genet. Genom. Med. 2018, 6, 261–267.
- Yang, Y.; Yang, C.; Zhu, Y.; Chen, H.; Zhao, R.; He, X.; Tao, L.; Wang, P.; Zhou, L.; Zhao, L.; et al. Intragenic and Extragenic Disruptions of FOXL2 Mapped by Whole Genome Low-Coverage Sequencing in Two BPES Families with Chromosome Reciprocal Translocation. Genomes 2014, 104, 170–176.
- Zahanova, S.; Meaney, B.; Łabieniec, B.; Verdin, H.; De Baere, E.; Nowaczyk, M.J. Blepharophimosis-Ptosis-Epicanthus Inversus Syndrome Plus. Clin. Dysmorphol. 2012, 21, 48–52.
- Zhang, L.; Wang, L.; Han, R.; Guan, L.; Fan, B.; Liu, M.; Ying, M.; Peng, H.; Li, N. Identification of the Forkhead Transcriptional Factor 2 (FOXL2) Gene Mutations in Four Chinese Families with Blepharophimosis Syndrome. Mol. Vis. 2013, 19, 2298–2305.
- Zhou, L.; Wang, J.; Wang, T. Functional Study on New FOXL2 Mutations Found in Chinese Patients with Blepharophimosis, Ptosis, Epicanthus Inversus Syndrome. BMC Med. Genet. 2018, 19, 121.
- Bunyan, D.J.; Thomas, N.S. Screening of a Large Cohort of Blepharophimosis, Ptosis, and Epicanthus Inversus Syndrome Patients Reveals a Very Strong PA-Ternal Inheritance Bias and a Wide Spectrum of Novel FOXL2 Mutations. Eur. J. Med. Genet. 2019, 62, 103668.
- Brocke, K.S.; Neu-Yilik, G.; Gehring, N.H.; Hentze, M.W.; Kulozik, A.E. The Human Intronless Melanocortin 4-Receptor Gene Is NMD Insensitive. Hum. Mol. Genet. 2002, 11, 331–335.
- Moumné, L.; Fellous, M.; Veitia, R.A. Deletions in the Polyalanine-Containing Transcription Factor FOXL2 Lead to Intranuclear Aggregation. Hum. Mol. Genet. 2005, 14, 3557–3564.
- De Baere, E.; Dixon, M.J.; Small, K.W.; Jabs, E.W.; Leroy, B.P.; Devriendt, K.; Gillerot, Y.; Mortier, G.; Meire, F.; Van Maldergem, L.; et al. Spectrum of FOXL2 Gene Mutations in Blepharophimosis-Ptosis-Epicanthus Inversus (BPES) Families Demonstrates a Genotype-Phenotype Correla-Tion. Hum. Mol. Genet. 2001, 10, 1591–1600.
- Brown, L.Y.; Brown, S.A. Alanine Tracts: The Expanding Story of Human Illness and Trinucleotide Repeats. Trends Genet. 2004, 20, 51–58.
- Kosaki, K.; Ogata, T.; Kosaki, R.; Sato, S.; Matsuo, N. A Novel Mutation in the FOXL2 Gene in a Patient with Blepharophimosis Syndrome: Differential Role of the Polyalanine Tract in the Development of the Ovary and the Eyelid. Ophthalmic Genet. 2002, 23, 43–47.
- Caburet, S.; Demarez, A.; Moumné, L.; Fellous, M.; De Baere, E.A.; Veitia, R. A Recurrent Polyalanine Expansion in the Transcription Factor FOXL2 Induces Extensive Nuclear and Cytoplasmic Protein Aggregation. J. Med. Genet. 2004, 41, 932–936.
- Moumné, L.; Dipietromaria, A.; Batista, F.; Kocer, A.; Fellous, M.; Pailhoux, E.; Veitia, R.A. Differential Aggregation and Functional Impairment Induced by Polyalanine Expansions in FOXL2, a Transcription Factor in-Volved in Cranio-Facial and Ovarian Development. Hum. Mol. Genet. 2007, 17, 1010–1019.
- Boccone, L.; Meloni, A.; Falchi, A.M.; Usai, V.; Cao, A. Blepharophimosis, Ptosis, Epicanthus Inversus Syndrome, a New Case Associated with de Novo Balanced Autosomal Translocation [46,XY,t(3;7)(q23;q32)]. Am. J. Med. Genet. 1994, 51, 258–259.
- Fukushima, Y.; Wakui, K.; Nishida, T.; Ueoka, Y. Blepharophimosis Sequence Andde Novo Balanced Autosomal Translocation [46, XY,t(3;4)(q23;p15.2)]: Possible Assignment of the Trait to 3q23. Am. J. Med. Genet. 1991, 40, 485–487.
- Kleinjan, D.-J.; Van Heyningen, V. Position Effect in Human Genetic Disease. Hum. Mol. Genet. 1998, 7, 1611–1618.
- Nikic, S.; Vaiman, D. Conserved Patterns of Gene Expression in Mice and Goats in the Vicinity of the Polled Intersex Syndrome (PIS) locus. Chromosom. Res. 2004, 12, 465–474.
- Pailhoux, E.; Vigier, B.; Schibler, L.; Cribiu, E.P.; Cotinot, C.; Vaiman, D. Positional Cloning of the PIS Mutation in Goats and Its Impact on Understanding Mammalian Sex-Differentiation. Genet. Sel. Evol. 2005, 37, S55.
- Uhlenhaut, N.H.; Treier, M. FOXL2 Function in Ovarian Development. Mol. Genet. Metab. 2006, 88, 225–234.
- Sills, E.S.; Alper, M.M.; Walsh, A.P. Ovarian Reserve Screening in Infertility: Practical Applications and Theoretical Directions for Research. Eur. J. Obstet. Gynecol. Reprod. Biol. 2009, 146, 30–36.
- Cohen, J.L.; Chabbert-Buffet, N.; Darai, E. Diminished Ovarian Reserve, Premature Ovarian Failure, Poor Ovarian Responder—A Plea for Universal Definitions. J. Assist. Reprod. Genet. 2015, 32, 1709–1712.
- Pastore, L.M.; Christianson, M.S.; Stelling, J.; Kearns, W.G.; Segars, J.H. Reproductive Ovarian Testing and the Alphabet Soup of Diagnoses: DOR, POI, POF, POR, and FOR. J. Assist. Reprod. Genet. 2018, 35, 17–23.
- Roth, L.; Alvero, R. Pregnancy in a Woman with Premature Ovarian Insufficiency Associated with Blepharo-phimosis, Ptosis, Epicanthus Inversus Syndrome Type I: A Case Report. Request PDF. J. Reprod. Med. 2014, 59, 87–89.




