
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Thomas Tu | + 1729 word(s) | 1729 | 2021-03-01 10:44:28 | | | |
| 2 | Peter Tang | Meta information modification | 1729 | 2021-03-10 10:34:05 | | |
Video Upload Options
Hepatitis B virus (HBV) is a globally-distributed pathogen and is a major cause of liver disease. HBV (or closely-related animal hepadnaviruses) can integrate into the host genome, but (unlike retroviruses) this integrated form is replication-defective.
1. Introduction
Chronic infection with the human hepatitis B virus (HBV) is one of the major drivers of liver disease and is the most common cause of liver cancer worldwide. Approximately one third of the world’s population has been exposed to the virus, and ~257 million people currently live with a chronic HBV infection [1][2]. Chronic HBV infections are usually life-long (with few exceptions) as the virus has developed several persistence mechanisms to escape immune surveillance, including replication via a highly-stable nuclear episomal template (cccDNA) mimicking a mini-chromosome. Another mode of escaping elimination by the immune systems is the expression of sub-viral particles that dampen antiviral immune responses by mechanisms not yet fully understood.
2. Natural History of Chronic Hepatitis B
Chronic HBV infections are generally asymptomatic for long periods (up to decades) as the virus replicates without triggering an antiviral immune response [3]. Decades later, a sub-optimal immune response is raised against HBV-expressing hepatocytes through yet-unknown mechanisms. The host response is usually inadequate to overcome viral persistence mechanisms, but instead leads to chronic inflammation, liver damage, and hepatic disease.
Surveillance and ongoing care for chronic HBV infection is complicated by psychosocial factors, including limited access to or distrust of the health care system, financial burdens, social stigma, and systemic discrimination [4]. A poor cascade of care results: only ~10% of cases are diagnosed and only ~3% are adequately treated even in some developed countries [5][6]. Thus, HBV-related disease generally progresses unmonitored and kills ~880,000 people annually through liver cirrhosis and liver cancer [1][2].
The mechanisms by which HBV causes liver cancer are still not well understood and are under intense investigation. One of the major factors thought to be involved in hepatocarcinogenesis is the integration of the viral DNA genome into cellular chromosomes. This reportedly induces pro-oncogenic pathways through several means (previously reviewed in detail [7][8][9]), including cis-mediated mechanisms (e.g., HBV DNA integrations modulating expression of proximal cellular genes) or trans-mediated mechanisms (e.g., chronic expression of particular viral antigens). This review focuses on how in vitro models have helped in furthering our understanding of how integrated HBV DNA contributes to virus persistence and pathogenesis.
3. HBV Structure
HBV is the prototypical member of the Hepadnaviridae family, which are small enveloped, hepatotropic DNA viruses (~3.2 kbp) replicating via reverse transcription. The double-stranded DNA genome of HBV can take two forms (Figure 1): relaxed circular DNA (rcDNA, generally present in ~90% of virions [10]) or double stranded linear DNA (dslDNA, ~10% of virions). Unlike the fully replication-competent relaxed circular form, the dslDNA-containing viral particles are replication-defective (detailed further below) but can integrate via non-homologous recombination into hepatocyte chromosomes. The HBV genome contains four overlapping open reading frames that encode for seven viral proteins, including: the HBV core antigen (HBcAg, which forms the viral capsid), e antigen (HBeAg, a secreted viral protein encoded by the core/pre-core ORF with reported tolerogenic properties [11]), surface protein (HBsAg, which has three forms large, medium and small—or LHBs, MHBs, and SHBs—that envelop the virion), polymerase (HBV pol, which is essential for viral reverse transcription and replication), and the x protein (HBx, which regulates transcription of viral genes from the cccDNA episome).
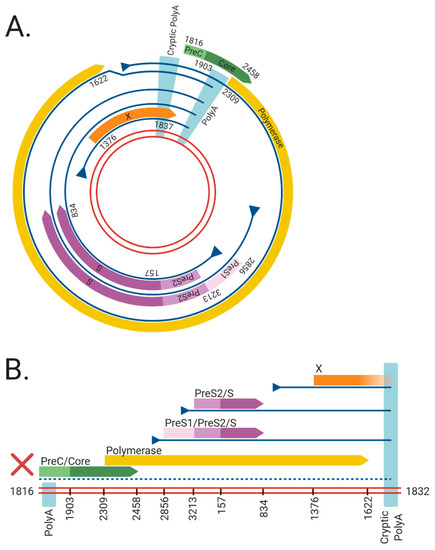
Figure 1. The viral RNAs and open reading frames (ORFs) expressed from the (A) cccDNA and (B) dslDNA forms of hepatitis B virus (HBV). The double red lines represent of the HBV DNA genome, blue lines represent viral mRNAs, triangles indicate transcriptional promotors, and coloured arrows represent viral ORFs. Nucleotide numbering of ORFs are shown as per Genbank Accession #AB241115. The viral mRNAs expressed from cccDNA terminate at a polyadenylation signal (poly A) located in the Core ORF. However, in dslDNA form, the canonical poly A signal is located at the 5′ end and mRNAs instead terminate at a non-canonical cryptic poly A signal. The mRNA coding for the HBV PreC/Core and Polymerase are separated from its promoter in the dslDNA form, leading to loss of their expression (dashed line). Figure generated using BioRender (https://biorender.com/).
4. HBV Replication
HBV replication starts with rcDNA-containing HBV particles attaching to heparan sulphate proteoglycans on the surface of hepatocytes [12][13][14] (Figure 2). This attachment sets up conditions for a high-affinity interaction between the preS-domain of LHBs and sodium taurocholate co-transporting polypeptide (NTCP), which acts as the functional receptor for HBV [15][16]. Binding is reportedly followed by clathrin-mediated endocytosis of the viral particle [17][18] and cytoplasmic release of the HBV nucleocapsid, which is subsequently transported to the nucleus and disassembles at the nuclear pore complex [19].

Figure 2. Replication cycle of hepatitis B virus (HBV) including integration into host genome. HBV virions attach to heparan sulphate proteoglycans (HSPG) on the cell surface, allowing for clathrin-mediated endocytosis via sodium taurochlorate co-transporting polypeptide (NTCP). Relaxed circular (rc)DNA genomes (top) are converted into covalently closed circular (ccc)DNA, which serve as the transcriptional template for viral mRNAs (vRNAs) and pregenomic (pg)RNA. Double stranded linear (dsl)DNA (bottom) can also form cccDNA but cannot code for functional rcDNA due to an additional 16nt insertion (asterisk) [20][21][22]. HBV pgRNA is encapsidated by viral capsid proteins and is reverse transcribed by the viral polymerase to produce either rcDNA or dslDNA. The mature nucleocapsids containing viral DNA are then enveloped by HBsAg embedded into host membranes and secreted as virions. As a secondary pathway, dslDNA can integrate into the host genome at double stranded DNA breaks, via non-homologous end joining. Integrated HBV DNA is replication-deficient due to rearrangements of the viral genome that abrogate expression of the viral polymerase and capsid proteins (Figure 1). Figure adapted from [23] and generated using BioRender (https://biorender.com/).
The HBV rcDNA genome is released into the nucleus, where it is repaired into the covalently closed circular DNA (cccDNA) form by multiple host DNA repair enzymes [24][25][26][27][28][29][30][31]. cccDNA acts as the template for all viral transcripts including pregenomic RNA (pgRNA), which is reverse-transcribed in nucleocapsids formed by the viral core antigen. This results in HBV DNA-containing nucleocapsids that are enveloped by host membranes studded with all three forms of HBsAg and then secreted from the cell via multi-vesicular bodies as virions.
5. HBV dslDNA and HBV DNA Integration
While the majority of intra-capsid reverse transcription events result in the formation of rcDNA, a minority of virions contain HBV dslDNA (though this ratio can fluctuate in patients in different stages of infection [10]). The production of dslDNA is dependent on binding and circularisation signals present on the reverse-transcribed strand of HBV DNA [32][33]. When dslDNA-containing virions infect a cell, HBV genome (instead of being converted into cccDNA) can integrate into cellular genome at the site of cellular double-stranded DNA breaks [34].
The role of integrated HBV DNA is currently unclear: unlike retroviruses, the integrated form of HBV is replication-deficient because of rearrangements that abrogate the expression of capsid, polymerase and functional pgRNA (Figure 1). Firstly, the orientation of the HBV dslDNA separates the HBcAg and pol at its 5′ end from their native promoter at its 3′ end. Moreover, the poly A signal shared by all viral transcripts is located on the 5′ end instead of the 3′ end of the dslDNA, leading to truncated transcripts lacking a canonical poly A. Most viral transcripts expressed from integrated HBV DNA can retain function by terminating using a non-canonical poly A signal retained at its 3′ end [35][36]. However, pgRNA is rendered non-functional as the premature termination removes structural elements from the 3′ end necessary for initiating pgRNA reverse transcription (namely, the direct repeat 1 and phi sequences [37][38][39][40]). Finally, terminal truncations on both ends of the integrated genome are introduced by the error-prone host DNA repair pathways during HBV DNA integration and can disrupt viral ORFs [34][41][42][43]. Together, these effects render integrated HBV DNA replication-deficient. No known cell lines with integrated HBV DNA derived from a natural infection produce infectious virus, though they can still express functional HBsAg [44][45][46][47].
Various functions have been ascribed to the integrated HBV DNA form (e.g., supporting viral persistence and pathogenesis). In vitro models (Table 1) have enabled detailed understanding of these aspects, including: the molecular mechanisms governing HBV DNA integration; its role as a source of HBsAg in a chronic infection; and how it is involved in HCC initiation and progression. The remainder of this review summarises the knowledge that these model systems have provided the field.
6. In Vitro Models of HBV DNA Integration
In vitro models vary in their ease of use (or reproducibility) in integration studies and inversely how closely they resemble integrations from a true infection in people with HBV. Tumour-derived cell lines from HBV patients contain integrations formed in the native setting but cannot be used to understand underlying mechanisms as they do not generate new integrations. Conversely, nuclear introduction of large numbers of HBV DNA molecules (via over-expressing constructs or transfection) can drive relatively high integration rates, allowing easy study of the integration process. However, this does not reflect a true infection; nuclear import of new HBV virions occurs relatively inefficiently in HBV infection [48][49][50][51], so levels of HBV DNA in infected hepatocytes is relatively low. Newly-developed infection models expressing the HBV receptor have solved this to some degree by largely recapitulating the virological aspects accurately. However, the low integration rate is challenging to detect without specialised methods, which can be difficult to reproduce. In essence, all studies need to be interpreted with these limitations in mind.
Table 1. A summary of in vitro model systems for human HBV DNA integration.
|
Model System |
Type |
Infectious HBV Produced? |
Forms New HBV DNA Integrations? |
Refs |
|---|---|---|---|---|
|
PLC/PRF/5 |
Tumour-derived cell line 1 |
No |
No |
|
|
Hep3B |
Tumour-derived cell line 1 |
No |
No |
|
|
HepG2.2.15 |
Engineered HBV-producer cell line 2 |
Yes |
Yes |
|
|
HepAD38 |
Engineered HBV-producer cell line 2 |
Yes |
Unknown |
[54] |
|
Transfection of HBV over-length constructs |
HBV transfection 3 |
Yes |
Unknown |
|
|
Transfection of HBV monomeric DNA |
HBV transfection 3 |
Yes |
Unknown |
|
|
Transfection of HBV virion DNA |
HBV transfection 3 |
Yes |
Yes |
[62] |
|
Transfection of in vitro transcribed HBV pgRNA |
HBV transfection 3 |
Yes |
Unknown |
[63] |
|
Huh7-NTCP |
HBV infection 4 |
Yes |
Yes |
|
|
HepG2-NTCP |
HBV infection 4 |
Yes |
Yes |
|
|
HepaRG |
HBV infection 4 |
Yes |
Yes |
|
|
HepaRG-NTCP |
HBV infection 4 |
Yes |
Yes |
|
|
Primary human hepatocytes |
HBV infection 4 |
Yes |
Yes |
1 Tumour-derived cell lines contain replication-deficient integrated HBV DNA acquired during infection. 2 Engineered HBV-producer cell lines contain stable recombinant replication-competent HBV constructs. 3 HBV transfection models involve ectopic introduction of viral DNA or RNA constructs. 4 HBV infection models involve bona fide viral infection of cells expressing the viral receptor NTCP.
References
- World Health Organization. Global Hepatitis Report 2017; WHO: Geneva, Switzerland, 2017.
- The Polaris Observatory Collaborators. Global Prevalence, Treatment, and Prevention of Hepatitis B Virus Infection in 2016: A Modelling Study. Lancet Gastroenterol. Hepatol. 2018, 3, 383–403.
- Mutz, P.; Metz, P.; Lempp, F.A.; Bender, S.; Qu, B.; Schöneweis, K.; Seitz, S.; Tu, T.; Restuccia, A.; Frankish, J.; et al. HBV Bypasses the Innate Immune Response and Does Not Protect HCV From Antiviral Activity of Interferon. Gastroenterology 2018, 154, 1791–1804.
- Tu, T.; Block, J.M.; Wang, S.; Cohen, C.; Douglas, M.W. The Lived Experience of Chronic Hepatitis B: A Broader View of Its Impacts and Why We Need a Cure. Viruses 2020, 12, 515.
- Allard, N.; MacLachlan, J.H.; Cowie, B.C. The cascade of care for Australians living with chronic hepatitis B: Measuring access to diagnosis, management and treatment. Aust. N. Z. J. Public Health 2015, 39, 255–259.
- Harris, A.M.; Osinubi, A.; Nelson, N.P.; Thompson, W.W. The hepatitis B care cascade using administrative claims data, 2016. Am. J. Manag. Care 2020, 26, 331–338.
- Tu, T.; Budzinska, M.A.; Shackel, N.A.; Jilbert, A.R. Conceptual models for the initiation of hepatitis B virus-associated hepatocellular carcinoma. Liver Int. 2015, 35, 1786–1800.
- Tu, T.; Bühler, S.; Bartenschlager, R. Chronic viral hepatitis and its association with liver cancer. Biol. Chem. 2017, 398, 817–837.
- Tu, T.; Budzinska, M.A.; Shackel, N.A.; Urban, S. HBV DNA Integration: Molecular Mechanisms and Clinical Implications. Viruses 2017, 9, 75.
- Zhao, X.-L.; Yang, J.-R.; Lin, S.-Z.; Ma, H.; Guo, F.; Yang, R.-F.; Zhang, H.-H.; Han, J.-C.; Wei, L.; Pan, X.-B. Serum viral duplex-linear DNA proportion increases with the progression of liver disease in patients infected with HBV. Gut 2015, 65, 502–511.
- Chen, M.S.; Billaud, J.-N.; Sällberg, M.; Guidotti, L.G.; Chisari, F.V.; Jones, J.; Hughes, J.; Milich, D.R. A function of the hepatitis B virus precore protein is to regulate the immune response to the core antigen. Proc. Natl. Acad. Sci. USA 2004, 101, 14913–14918.
- Schulze, A.; Gripon, P.; Urban, S. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology 2007, 46, 1759–1768.
- Leistner, C.M.; Gruen-Bernhard, S.; Glebe, D. Role of glycosaminoglycans for binding and infection of hepatitis B virus. Cell. Microbiol. 2007, 10, 122–133.
- Sureau, C.; Salisse, J. A conformational heparan sulfate binding site essential to infectivity overlaps with the conserved hepatitis B virus A-determinant. Hepatology 2012, 57, 985–994.
- Ni, Y.; Lempp, F.A.; Mehrle, S.; Nkongolo, S.; Kaufman, C.; Fälth, M.; Stindt, J.; Königer, C.; Nassal, M.; Kubitz, R.; et al. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 2014, 146, 1070–1083.
- Yan, H.; Zhong, G.; Xu, G.; He, W.; Jing, Z.; Gao, Z.; Huang, Y.; Qi, Y.; Peng, B.; Wang, H.; et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 2012, 1, e00049.
- Herrscher, C.; Pastor, F.; Burlaud-Gaillard, J.; Dumans, A.; Seigneuret, F.; Moreau, A.; Patient, R.; Eymieux, S.; De Rocquigny, H.; Hourioux, C.; et al. Hepatitis B virus entry into HepG2-NTCP cells requires clathrin-mediated endocytosis. Cell. Microbiol. 2020, 22, e13205.
- Umetsu, T.; Inoue, J.; Kogure, T.; Kakazu, E.; Ninomiya, M.; Iwata, T.; Takai, S.; Nakamura, T.; Sano, A.; Shimosegawa, T. Inhibitory effect of silibinin on hepatitis B virus entry. Biochem. Biophys. Rep. 2018, 14, 20–25.
- Rabe, B.; Delaleau, M.; Bischof, A.; Foss, M.; Sominskaya, I.; Pumpens, P.; Cazenave, C.; Castroviejo, M.; Kann, M. Nuclear Entry of Hepatitis B Virus Capsids Involves Disintegration to Protein Dimers followed by Nuclear Reassociation to Capsids. PLoS Pathog. 2009, 5, e1000563.
- Yang, W.; Summers, J. Illegitimate replication of linear hepadnavirus DNA through nonhomologous recombination. J. Virol. 1995, 69, 4029–4036.
- Yang, W.; Mason, W.S.; Summers, J. Covalently closed circular viral DNA formed from two types of linear DNA in woodchuck hepatitis virus-infected liver. J. Virol. 1996, 70, 4567–4575.
- Yang, W.; Summers, J. Infection of ducklings with virus particles containing linear double-stranded duck hepatitis B virus DNA: Illegitimate replication and reversion. J. Virol. 1998, 72, 8710–8717.
- Budzinska, M.A.; Shackel, N.A.; Urban, S.; Tu, T. Sequence analysis of integrated hepatitis B virus DNA during HBeAg-seroconversion. Emerg. Microbes Infect. 2018, 7, 142.
- Wei, L.; Ploss, A. Core components of DNA lagging strand synthesis machinery are essential for hepatitis B virus cccDNA for-mation. Nat. Microbiol. 2020, 5, 715–726.
- Kitamura, K.; Que, L.; Shimadu, M.; Koura, M.; Ishihara, Y.; Wakae, K.; Nakamura, T.; Watashi, K.; Wakita, T.; Muramatsu, M. Flap endonuclease 1 is involved in cccDNA formation in the hepatitis B virus. PLoS Pathog. 2018, 14, e1007124.
- Tang, L.; Sheraz, M.; McGrane, M.; Chang, J.; Guo, J.-T. DNA Polymerase alpha is essential for intracellular amplification of hepatitis B virus covalently closed circular DNA. PLoS Pathog. 2019, 15, e1007742.
- Qi, Y.; Gao, Z.; Xu, G.; Peng, B.; Liu, C.; Yan, H.; Yao, Q.; Sun, G.; Liu, Y.; Tang, D.; et al. DNA Polymerase kappa Is a Key Cellular Factor for the Formation of Covalently Closed Circular DNA of Hepatitis B Virus. PLoS Pathog. 2016, 12, e1005893.
- Long, Q.; Yan, R.; Hu, J.; Cai, D.; Mitra, B.; Kim, E.S.; Marchetti, A.; Zhang, H.; Wang, S.; Liu, Y.; et al. The role of host DNA ligases in hepadnavirus covalently closed circular DNA formation. PLoS Pathog. 2017, 13, e1006784.
- Sheraz, M.; Cheng, J.; Tang, L.; Chang, J.; Guo, J.-T. Cellular DNA Topoisomerases Are Required for the Synthesis of Hepatitis B Virus Covalently Closed Circular DNA. J. Virol. 2019, 93.
- Königer, C.; Wingert, I.; Marsmann, M.; Rösler, C.; Beck, J.; Nassal, M. Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis B viruses. Proc. Natl. Acad. Sci. USA 2014, 111, E4244–E4253.
- Cui, X.; McAllister, R.; Boregowda, R.; Sohn, J.A.; Ledesma, F.C.; Caldecott, K.W.; Seeger, C.; Hu, J. Does Tyrosyl DNA Phosphodiesterase-2 Play a Role in Hepatitis B Virus Genome Repair? PLoS ONE 2015, 10, e0128401.
- Haines, K.M.; Loeb, D.D. The Sequence of the RNA Primer and the DNA Template Influence the Initiation of Plus-strand DNA Synthesis in Hepatitis B Virus. J. Mol. Biol. 2007, 370, 471–480.
- Lewellyn, E.B.; Loeb, D.D. Base Pairing between cis-Acting Sequences Contributes to Template Switching during Plus-Strand DNA Synthesis in Human Hepatitis B Virus. J. Virol. 2007, 81, 6207–6215.
- Yang, W.; Summers, J. Integration of hepadnavirus DNA in infected liver: Evidence for a linear precursor. J. Virol. 1999, 73, 9710–9717.
- Schütz, T.; Kairat, A.; Schröder, C.H. DNA Sequence Requirements for the Activation of a CATAAA Polyadenylation Signal within the Hepatitis B Virus X Reading Frame: Rapid Detection of Truncated Transcripts. Virology 1996, 223, 401–405.
- Wooddell, C.I.; Lik-Yuen, C.H.; Chan, H.L.-Y.; Gish, R.G.; Locarnini, S.A.; Chavez, D.; Ferrari, C.; Given, B.D.; Hamilton, J.; Kanner, S.B.; et al. RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci. Transl. Med. 2017, 9, eaan0241.
- Liu, N.; Ji, L.; Maguire, M.L.; Loeb, D.D. cis-Acting Sequences That Contribute to the Synthesis of Relaxed-Circular DNA of Human Hepatitis B Virus. J. Virol. 2004, 78, 716–723.
- Wang, G.H.; Seeger, C. Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol. 1993, 67, 6507–6512.
- Oropeza, C.E.; McLachlan, A. Complementarity between epsilon and phi sequences in pregenomic RNA influences hepatitis B virus replication efficiency. Virology 2007, 359, 371–381.
- Nassal, M.; Rieger, A. A bulged region of the hepatitis B virus RNA encapsidation signal contains the replication origin for dis-continuous first-strand DNA synthesis. J. Virol. 1996, 70, 2764–2773.
- Bill, C.A.; Summers, J. Genomic DNA double-strand breaks are targets for hepadnaviral DNA integration. Proc. Natl. Acad. Sci. USA 2004, 101, 11135–11140.
- Summers, J.; Mason, W.S. Residual integrated viral DNA after hepadnavirus clearance by nucleoside analog therapy. Proc. Natl. Acad. Sci. USA 2003, 101, 638–640.
- Mason, W.S.; Liu, C.; Aldrich, C.E.; Litwin, S.; Yeh, M.M. Clonal Expansion of Normal-Appearing Human Hepatocytes during Chronic Hepatitis B Virus Infection. J. Virol. 2010, 84, 8308–8315.
- Alexander, J.; Bey, E.; Whitcutt, J.M.; Gear, J.H. Adaptation of cells derived from human malignant tumours to growth In Vitro. S. Afr. J. Med Sci. 1976, 41, 89–98.
- Dejean, A.; Bréchot, C.; Tiollais, P.; Wain-Hobson, S. Characterization of integrated hepatitis B viral DNA cloned from a human hepatoma and the hepatoma-derived cell line PLC/PRF/5. Proc. Natl. Acad. Sci. USA 1983, 80, 2505–2509.
- Aden, D.P.; Fogel, A.; Plotkin, S.; Damjanov, I.; Knowles, B.B. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nat. Cell Biol. 1979, 282, 615–616.
- Knowles, B.B.; Howe, C.C.; Aden, D.P. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science 1980, 209, 497–499.
- Tu, T.; Budzinska, M.A.; Vondran, F.W.R.; Shackel, N.A.; Urban, S. Hepatitis B virus DNA integration occurs early in the viral life cycle in an In Vitro infection model via NTCP-dependent uptake of enveloped virus particles. J. Virol. 2018.
- Volz, T.; Allweiss, L.; Ben Ḿbarek, M.; Warlich, M.; Lohse, A.W.; Pollok, J.M.; Alexandrov, A.; Urban, S.; Petersen, J.; Lütgehetmann, M.; et al. The entry inhibitor Myrcludex-B efficiently blocks intrahepatic virus spreading in humanized mice previously infected with hepatitis B virus. J. Hepatol. 2013, 58, 861–867.
- Allweiss, L.; Volz, T.; Giersch, K.; Kah, J.; Raffa, G.; Petersen, J.; Lohse, A.W.; Beninati, C.; Pollicino, T.; Urban, S.; et al. Proliferation of primary human hepatocytes and prevention of hepatitis B virus reinfection efficiently deplete nuclear cccDNA In Vivo. Gut 2018, 67, 542–552.
- Tu, T.; Zehnder, B.; Qu, B.; Urban, S. De novo synthesis of Hepatitis B virus nucleocapsids is dispensable for the maintenance and transcriptional regulation of cccDNA. JHEP Rep. 2020.
- Dandri, M.; Burda, M.R.; Bürkle, A.; Zuckerman, D.M.; Will, H.; Rogler, C.E.; Greten, H.; Petersen, J. Increase in de novo HBV DNA integrations in response to oxidative DNA damage or inhibition of poly (ADP-ribosyl) ation. Hepatology 2002, 35, 217–223.
- Ruan, P.; Dai, X.; Sun, J.; He, C.; Huang, C.; Zhou, R.; Chemin, I. Integration of hepatitis B virus DNA into p21-activated kinase 3 (PAK3) gene in HepG2.2.15 cells. Virus Genes 2020, 56, 168–173.
- Ladner, S.K.; Otto, M.J.; Barker, C.S.; Zaifert, K.; Wang, G.H.; Guo, J.T.; Seeger, C.; King, R.W. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: A novel system for screening potential inhibitors of HBV replication. Antimicrob. Agents Chemother. 1997, 41, 1715–1720.
- Sureau, C.; Romet-Lemonne, J.-L.; Mullins, J.I.; Essex, M. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell 1986, 47, 37–47.
- Sureau, C.; Eichberg, J.W.; Hubbard, G.B.; Romet-Lemonne, J.L.; Essex, M. A molecularly cloned hepatitis B virus produced In Vitro is infectious in a chimpanzee. J. Virol. 1988, 62, 3064–3067.
- Sells, M.A.; Zelent, A.Z.; Shvartsman, M.; Acs, G. Replicative intermediates of hepatitis B virus in HepG2 cells that produce infectious virions. J. Virol. 1988, 62, 2836–2844.
- Acs, G.; Sells, M.A.; Purcell, R.H.; Price, P.; Engle, R.; Shapiro, M.; Popper, H. Hepatitis B virus produced by transfected Hep G2 cells causes hepatitis in chimpanzees. Proc. Natl. Acad. Sci. USA 1987, 84, 4641–4644.
- Will, H.; Cattaneo, R.; Koch, H.-G.; Darai, G.; Schaller, H.; Schellekens, H.; Van Eerd, P.M.C.A.; Deinhardt, F. Cloned HBV DNA causes hepatitis in chimpanzees. Nat. Cell Biol. 1982, 299, 740–742.
- Guo, X.; Chen, P.; Hou, X.; Xu, W.; Wang, D.; Wang, T.-Y.; Zhang, L.; Zheng, G.; Gao, Z.-L.; He, C.-Y.; et al. The recombined cccDNA produced using minicircle technology mimicked HBV genome in structure and function closely. Sci. Rep. 2016, 6, 25552.
- Gunther, S.; Sommer, G.; Von Breunig, F.; Iwanska, A.; Kalinina, T.; Sterneck, M.; Will, H. Amplification of full-length hepatitis B virus genomes from samples from patients with low levels of viremia: Fre-quency and functional consequences of PCR-introduced mutations. J. Clin. Microbiol. 1998, 36, 531–538.
- Tu, T.; Zehnder, B.; Levy, M.; Micali, G.; Tran, L.; Dabere, O.; Main, N.; Shackel, N.; Urban, S. Hepatitis B virus (HBV) DNA integration is not driven by viral proteins. Jahrestag. Dtsch. Arb. Stud. Leb. 2019, 57.
- Yu, Y.; Schneider, W.M.; Michailidis, E.; Acevedo, A.; Ni, Y.; Ambrose, C.; Zou, P.; Kabbani, M.; Quirk, C.; Jahan, C.; et al. An RNA-based system to study hepatitis B virus replication and select drug-resistance mutations. bioRxiv 2019.




