
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Xavier Salom-Roig | + 1154 word(s) | 1154 | 2021-02-19 07:44:53 | | | |
| 2 | Camila Xu | Meta information modification | 1154 | 2021-03-18 03:44:17 | | |
Video Upload Options
Aurilides are a class of depsipeptides occurring mainly in marine cyanobacteria. Members of the aurilide family have shown to exhibit strong cytotoxicity against various cancer cell lines. These compounds bear a pentapeptide, a polyketide, and an α-hydroxy ester subunit in their structure.
1. Introduction
Since the late 1960s, marine organisms have been intensively explored as hopeful resources for anticancer drugs, and a diverse array of new compounds are still being discovered every year. In particular, the sea hare Dolabella auricularia is known as a prolific producer of cytotoxic and/or antitumor structurally unique secondary metabolites such as dolastatins 10 and 15 [1]. From a specimen of this marine organism collected in the Japanese sea, Suenaga et al. [2] isolated in 1996 aurilide (1), a 26-membered cyclodepsipeptide, which exhibits a strong cytotoxicity against HeLa S3 cells with an IC50 of 0.011 μg/mL.
In the next two decades, the aurilide family expanded with the discovery of ten new members, which were structurally analogous to aurilide (structures of this family are shown in Figure 1).
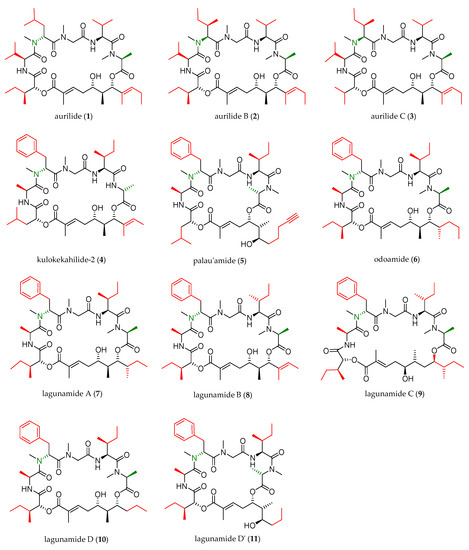
Figure 1. Aurilide family members.
As shown in Table 1, cyanobacterium Lyngbya majuscule was the source of aurilide B (2) and C (3), and cephalaspidean mollusc Philinopsis speciosa furnished kulokekahilide-2 (4). Palau’amide (5), odoamide (6), and lagunamides A (7), B (8), C (9), D (10), and D’ (11) were isolated from different sources of marine cyanobacteria.
Table 1. Marine sources of the aurilide family members.
| Aurilide-Family Member | Year of Isolation | Marine Source | Collection Site |
|---|---|---|---|
| aurilide (1) | 1996 [2] | sea hare Dolabella auricularia | Japanese sea |
| aurilide B (2) | 2006 [3] | cyanobacterium Lyngbya majuscula | Papua New Guinea |
| aurilide C (3) | 2006 [3] | cyanobacterium Lyngbya majuscula | Papua New Guinea |
| kulokekahilide-2 (4) | 2004 [4] | cephalaspidean MolluskPhilinopsis speciosa | |
| palau’amide (5) | 2000 [5] | cyanobacterium Lyngbya (Oscillatoriaceae) | Ulong Channel, Palau |
| odoamide (6) | 2016 [6] | cyanobacterium Okeania sp. | Japanese sea |
| lagunamide A (7) | 2010 [7] | cyanobacterium Lyngbya majuscula | western lagoon of Pulau Hantu Besar, Singapore |
| lagunamide B (8) | 2010 [7] | cyanobacterium Lyngbya majuscula | western lagoon of Pulau Hantu Besar, Singapore |
| lagunamide C (9) | 2011 [8] | cyanobacterium Lyngbya majuscula | western lagoon of Pulau Hantu Besar, Singapore |
| lagunamide D (10) and D’(11) | 2019 [9] | collection of marine cyanobacteria (a mixture of Dichothrix sp. and Lyngbyasp. in a ratio of 1:1 with minor amount of Rivularia sp. present) | Loggerhead Key in the Dry Tortugas in Florida |
Since the first synthesis of aurilide in 1996 by Suenaga and co-workers [2], the aurilide family has attracted considerable attention from the synthetic community due to potent antiproliferative activity as well as synthetically challenging molecular architecture and additional total syntheses of aurilide and other members of the aurilide class have been reported.
2. Structural Features
Structurally, the aurilide family members can be described as cyclic depsipeptides, whose framework can be divided into three subunits: an α-hydroxy acid residue, a polyketide segment containing three or four stereogenic centers, and a pentapeptide.
The size of the macrocycle differs slightly; eight of them (1−4, 6−8 and 10) are 26-membered rings, and lagunamide C (9) is a 27-membered ring, while plau’amide (5) and lagunamide D’ (11) are 24-membered rings.
2.1. Differences in the α-Hydroxy Acid and the Polyketide Subunits
Three different α-hydroxy acids (Figure 2) can be found in the macrocyclic structure of the aurilide family members. 2-Hydroxyisoleucic acid (Hila) is the most common residue and can be found in aurilide (1), aurilide B (2), lagunamides A (7), B (8), C (9), D (10), and D’ (11) and odoamide (6). Kulokekahilide-2 (4) and palau’amide (5) bear a 2-hydroxyisocaproic acid (Hica), whereas 2-hydroxyisovaleric acid (Hiva) is exclusive to aurilide C (3).
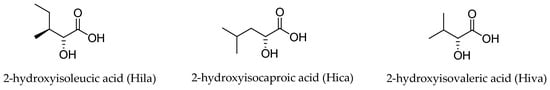
Figure 2. The three α-hydroxy acids present in the structure of the aurilide family members.
The polyketide subunit consists of an α,β-unsaturated 5,7-dihydroxy acid bearing three contiguous stereocenters at C5, C6, and C7 with the relative configuration 5,6- and 6,7- anti and an aliphatic saturated or unsaturated side-chain at C8, whose nature is depending on the member of the family (Figure 3). Odoamide (6) and lagunamide A (7) bear a fourth stereocenter in the side chain, whose absolute configuration is R for the first compound and S for the second one. Lagunamide C (9) can be considered as a subclass of aurilide-related compounds that has a ring expansion due to the additional methylene carbon inserted in the polyketide-derived moiety at C7.
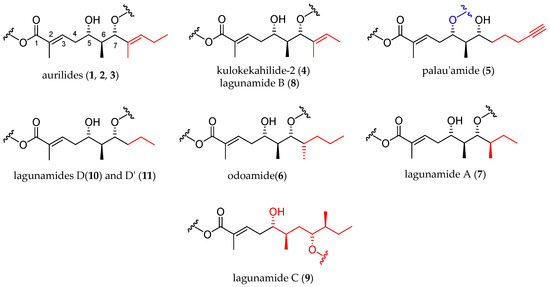
Figure 3. Polyketide moieties of the aurilide family members.
2.2. Differences in the Pentapeptide Fragment
Most of the depsipeptides of the aurilide class share similar peptide substructures. However, some of them have unique structural motifs and particularly in aurilides, three of the amino acids have different side chains. The first amino acid (AA1, Table 2) is l-N-methylalanine with the sole exception of kulokekalide-2 (4) in whose structure the other enantiomer is present. The third amino acid (AA3) is in all cases sarcosine, and the fifth one (AA5) is l-alanine, except for aurilides (1−3), which bear an l-valine. The main differences concern the second and fourth amino acid. Concerning the second amino acid (AA2), l-valine is exclusive to aurilides (1−3), while l-isoleucine is present in the other members of the family, although the absolute configuration of the side-chain stereocenter is not always the same (usually S but R in two cases). The fourth amino acid (AA4) normally found is d-phenylalanine, but in the case of aurilide (1), AA4 is replaced by l-leucine and in aurilides B (2) and C (3), it is replaced by d-isoleucine.
Table 2. Pentapeptide side chains in the different members of the aurilide family.
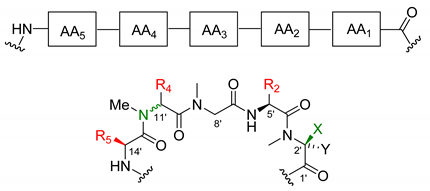
| R5 | R4 | R2 | C11′ Configuration | X | Y | |
|---|---|---|---|---|---|---|
| Aurilide (1) | i-Pr | i-Bu | i-Pr | R | Me | H |
| Aurilide B (2) | i-Pr | (R)-sec-butyl | i-Pr | S | Me | H |
| Aurilide C (3) | i-Pr | (R)-sec-butyl | i-Pr | S | Me | H |
| Kulokekahilide-2 (4) | Me | Bn | (S)-sec-butyl | R | H | Me |
| Palau’amide (5) | Me | Bn | (S)-sec-butyl | R | Me | H |
| Odoamide (6) | Me | Bn | (S)-sec-butyl | R | Me | H |
| Lagunamide A (7) | Me | Bn | (S)-sec-butyl | R | Me | H |
| Lagunamide B (8) | Me | Bn | (R)-sec-butyl | R | Me | H |
| Lagunamide C (9) | Me | Bn | (R)-sec-butyl | R | Me | H |
| Lagunamide D (10) | Me | Bn | (S)-sec-butyl | R | Me | H |
| Lagunamide D’ (11) | Me | Bn | (S)-sec-butyl | R | Me | H |
3. Biological Activities
Different members of the aurilide family exhibit strong cytotoxicity against various cancer cell lines, often in the range of nM concentrations, as summarized in Table 3.
Table 3. Cytotoxic activities of the natural aurilide class members (IC50 values in nM, *(LC50) in nM).
| HeLaS3 | P388 | BJ | BJ Shp 53 |
PC 3 |
SK-OV-3 | HCT8 | NCI-H460 | Neuro-2a * | MDA-MB-435 | A-10 | KB | A549 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aurilide (1) [2] | 11 | ||||||||||||
| Aurilide B (2) [3] | 10 | 40 | |||||||||||
| Aurilide C (3) [3] | 50 | 130 | |||||||||||
| Kulokekalide-2 (4) [4] | 4.2 | 7.5 | 14.6 | 59.1 | |||||||||
| Palau’amide (5) [5] | 13 | ||||||||||||
| Odoamide (6) [6][10] | 26.3 | 4.2 | |||||||||||
| Lagunamide A (7) [7][11] | 6.4 | 20.2 | 58.8 | 2.5 | 3.8 | 1.6 | 2.9 | ||||||
| Lagunamide B (8) [7][11] | 20.5 | 5.2 | |||||||||||
| Lagunamide C (9) [8] | 24.4 | 2.6 | 4.5 | 2.1 | 2.4 | ||||||||
| Lagunamide D (10) [9] | 7.1 | ||||||||||||
| Lagunamide D’(11) [9] | 68.2 |
Aurilide has been identified as an inducer of apoptosis by interfering with the morphogenesis of mitochondria [12]. Investigation of the mechanism of cytotoxicity showed that aurilide (1) binds to prohibitin 1 (PHB1), a mitochondria inner membrane protein, which in turn activates the proteolytic processing of optic atrophy 1 (OPA1), leading to mitochondrial fragmentation and apoptosis [13].
Lagunamide A induces caspase-mediated mitochondrial apoptosis in A549 cells [14]. Indeed, lagunamide A causes mitochondrial dysfunction followed by cell death along with the dissipation of mitochondrial membrane potential (Δφm) and overproduction of reactive oxygen species (ROS). It was proved that both anti- and pro-apoptotic B-cell lymphoma 2 (Bcl-2) family proteins, especially myeloid cell leukemia-1 (Mcl-1), participated in lagunamide A-induced mitochondrial apoptosis. The overexpression of Mcl-1 partly rescued A549 cells from lagunamide A-induced apoptosis.
Moreover, lagunamides A (7), B (8), and C (9) displayed significant antimalarial properties, with IC50 values of 0.19, 0.91, and 0.29 μM, respectively, when tested against Plasmodium falciparum [7].
References
- Pettit, G.R.; Kamano, Y.; Herald, C.L.; Fujii, Y.; Kizu, H.; Boyd, M.R.; Boettner, F.E.; Doubek, D.L.; Schmidt, J.M.; Chapuis, J.-C.; et al. Isolation of dolastatins 10–15 from the marine mollusc dolabella auricularia. Tetrahedron Lett. 1993, 49, 9151–9170.
- Suenaga, K.; Tsuyoshi, M.; Shibata, T.; Itoh, T.; Kigoshi, H.; Yamada, H. Isolation and stereostructure of aurilide, a novel cyclodepsipeptide from the Japanese sea hare Dolabella auricularia. Tetrahedron Lett. 1996, 37, 6771–6774.
- Han, B.; Gross, H.; Goeger, D.E.; Mooberry, S.L.; Gerwick, W.H. Aurilides B and C, cancer cell toxins from a Papua New Guinea collection of the marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2006, 69, 572–575.
- Nakao, Y.; Yoshida, W.Y.; Takada, Y.; Kimura, J.; Yang, L.; Mooberry, S.L.; Scheuer, P.J. Kulokekahilide-2, a cytotoxic depsipeptide from a cephalaspidean mollusk Philinopsis speciosa. J. Nat. Prod. 2004, 67, 1332–1340.
- Williams, P.G.; Yoshida, W.Y.; Quon, M.K.; Moore, R.E.; Paul, V.J. The structure of Palau’amide, a potent cytotoxin from a species of the marine cyanobacterium Lyngbya. J. Nat. Prod. 2003, 66, 1545–1549.
- Sueyoshi, K.; Kaneda, M.; Sumimoto, S.; Oishi, S.; Fujii, N.; Suenaga, K.; Teruya, T. Odoamide, a cytotoxic cyclodepsipeptide from the marine cyanobacterium Okeania sp. Tetrahedron 2016, 72, 5472–5478.
- Tripathi, A.; Puddick, J.; Prinsep, M.R.; Rottmann, M.; Tan, L.T. Lagunamides A and B: Cytotoxic and antimalarial cyclodepsipeptides from the marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2010, 73, 1810–1814.
- Tripathi, A.; Puddick, J.; Prinsep, M.R.; Rottmann, M.; Chan, K.P.; Chen, D.Y.-K.; Tan, L.T. Lagunamide C, a cytotoxic cyclodepsipeptide from the marine cyanobacterium Lyngbya majuscula. Phytochemistry 2011, 72, 2369–2375.
- Luo, D.; Putra, M.Y.; Ye, T.; Paul, V.J.; Luesch, H. Isolation, structure elucidation and biological evaluation of lagunamide D: A new cytotoxic macrocyclic depsipeptide from marine cyanobacteria. Mar. Drugs 2019, 17, 83.
- Kaneda, M.; Kawaguchi, S.; Fujii, N.; Ohno, H.; Oishi, S. Structure−activity relationship study on odoamide: Insights into the bioactivities of aurilide-family hybrid peptide−polyketides. ACS Med. Chem. Lett. 2018, 9, 365–369.
- Tripathi, A.; Fang, W.; Leong, D.T.; Tan, L.T. Biochemical Studies of the lagunamides, potent cytotoxic cyclic depsipeptides from the marine cyanobacterium Lyngbya majuscula. Mar. Drugs 2012, 10, 1126–1137.
- Semenzato, M.; Cogliati, S.; Scorrano, L. Prohibitin(g) cancer: Aurilide and killing by opa1-dependent cristae remodeling. Chem. Biol. 2011, 18, 8–9.
- Sato, S.; Murata, A.; Orihara, T.; Shirakawa, T.; Suenaga, K.; Kigoshi, H.; Uesugi, M. Marine natural product aurilide activates the OPA1-mediated apoptosis by binding to prohibitin. Chem. Biol. 2011, 18, 131–139.
- Huang, X.; Huang, W.; Li, L.; Sun, X.; Song, S.; Xu, Q.; Zhang, L.; Wei, B.-G.; Deng, X. Structure determinants of lagunamide A for anticancer activity and its molecular mechanism of mitochondrial apoptosis. Mol. Pharmaceutics. 2016, 13, 3756–3763.





