
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Delia Delia Mare | + 2398 word(s) | 2398 | 2021-02-24 07:24:40 | | | |
| 2 | Camila Xu | Meta information modification | 2398 | 2021-03-03 10:40:53 | | |
Video Upload Options
E. coli is a versatile microorganism, and even if the invasive infections are those that are more likely to evolve with life threatening complications, the diarrheagenic strains of E. coli are still important pathogens, especially in the pediatric departments.
1. Introduction
Diarrheal disease is still a major health problem, and it is considered an important cause of morbidity and mortality, especially in infants and children under five years old. Although the disease primarily affects populations from developing countries, where sanitation, water supplies, and the medical addressability rate are inadequate, there are still reported cases in developed countries [1][2][3][4]. The etiology of the infectious diarrheal disease is influenced by the geographic area. Viruses (such as rotavirus, adenovirus, norovirus) are the most frequent etiological agents worldwide. Infections with Campylobacter spp. and Salmonella spp. are frequently reported in the developed countries, while diarrheagenic strains of E. coli, Vibrio spp. are encountered mostly in developing countries [5][6][7].
In 1885, Theodor Escherich, a German pediatrician described, for the first time, Bacterium coli commune, as a commensal Gram-negative rod from the healthy individual’s intestinal flora. These rods were afterward renamed Escherichia coli (E. coli), in his honor. Only after a few decades, in 1935, they were considered pathogens, being associated with an outbreak of “cholera infantum”, a severe gastroenteritis affecting neonates [8][9]. From that moment, a remarkable number of studies were conducted, describing E. coli from numerous points of view (serological, antibiotic resistance, molecular, genetic engineering, sequencing, etc.), leading to the fact that, in our days, it is considered as one of the best-characterized bacteria. Even so, there are still many unknowns about this pathogen, which, even nowadays, is responsible for a large number of infections in healthy or immunocompromised persons (diarrhea, pneumonia, urinary, wounds infections, sepsis, meningitidis) [10][11].
E. coli is a versatile microorganism, and even if the invasive infections are those that are more likely to evolve with life threatening complications, the diarrheagenic strains of E. coli are still important pathogens, especially in the pediatric departments. Among these strains, Enteropathogenic E. coli (EPEC) is a particularly important one, as it is a bacterial etiological agent in a pathology that is dominated by viruses, making it easy to be “forgotten” or not taken into consideration, especially in the case of infantile diarrhea. The fact that these strains present multiple phenotypic differences, that they possess different, multiple virulence factors (some of them belonging to other pathotypes), and that they are still evolving, makes them a group of bacteria that will always be of interest for physicians from different medical departments. The main objective of this review was to summarize the most important, relevant, and up to date information available in the literature about this reemerging pathogen, and to provide an overview of the most important characteristics of EPEC strains.
The diarrheagenic strains of E. coli are a very homogeneous group of intestinal pathogens, with different characteristics regarding the antigen structure, virulence mechanisms, host colonization sites, and clinical evolution [11]. The O (lipopolysaccharide) and H (flagellar) antigens are used for the serological classification of the pathogenic strain of E. coli. Because of the high diversity of these antigens (187 O antigens, 53 H antigens), currently, serotyping is considered a laborious, costly, and unreliable diagnostic tool due to the cross-reactivity between different serogroups [12][13]. Most of the recent diagnostic protocols recommend the classification/identification of diarrheagenic E. coli using molecular methods, which can identify the virulence factors of these strains. Based on the presence/absence of virulence factors, the diarrheagenic strains of E. coli are classified into 6 major pathotypes and a hybrid one (Table 1). The horizontal gene transfer between different strains is an important mechanism that leads to the diversity of these strains, keeping them in a constant movement between the classically described pathotypes. Some strains that combine virulence factors from different pathotypes are considered hybrids, and they can be potentially more virulent than the original strains [6][11][14]. Plasmids, bacteriophages, transposons, pathogenicity islands, insertion sequences (mobile genetic elements–MGEs) plays important roles in the genomic plasticity of E. coli [15]. Whole genome sequencing is considered the gold standard technique which can accurately serotype strains and evaluate relatedness among isolates based on differences in gene content or allelic variation [16][17][18][19]. This technique revealed that E. coli presents a conserved core of genes (common to pathogenic and commensal strains as well) that acquired, by horizontal gene transfer, small cluster of genes and genomic islands associated with virulence [20].
Table 1. Pathotypes of Diarrheagenic E. coli.
| Pathotype of Diarrheagenic E. coli | Acronym | Virulence Factors | Diagnostic Targets for PCR | Disease | Clinical Symptoms | Ref |
|---|---|---|---|---|---|---|
| Enteropathogenic E. coli |
EPEC | Locus of enterocyte effacement (LEE), intimin, bundle-forming pilus | bae, bfpA | Acute/persistent diarrhea in children | Watery diarrhea, vomiting | [6][17][21] |
| Enterohaemorrhagic E. coli (Shiga toxin-producing) |
EHEC/STEC | Shiga toxin 1 and/or 2 LEE, adhesins (EHEC) |
stx1, stx2, eae, ehxA, bfp | Hemolytic-uremic syndrome, hemorrhagic colitis | Bloody diarrhea | [6][11][17] |
| Enteroinvasive E. coli |
EIEC | Shiga toxin, hemolysin, cellular invasion, Ipa | ipaH, other ipa genes | Shigellosis-like syndrome | Watery, bloody diarrhea | [6][11] |
| Adherent Invasive E. coli |
AIEC | Type 1 fimbriae, cellular invasion | none | Associated with Crohn disease | Persistent intestinal inflammation | [12][17][22] |
| Enterotoxigenic E. coli |
ETEC | Heat-labile and heat-stable toxins, CFAs (colonization factors) | elt, est | Traveler’s diarrhea | Watery diarrhea, vomiting | [6][11][17] |
| Diffusely Adherent E. coli |
DAEC | Adhesins | Afa/Dr adhesins | Diarrhea in children | Watery diarrhea | [6][17][23] |
| Enteroaggregative E. coli (hybrid pathotype) |
EAEC | Adhesins, toxins and secreted proteins | pAA, AggR, AAFs | Diarrhea in children | Vomiting diarrhea (with mucus) | [6][11] |
2. EPEC Definition and Classification
EPEC was the first identified pathotype of diarrheagenic E. coli, being the causative agent of a few outbreaks of infantile diarrhea in 1940–1950 [24]. Until a few decades ago, the O (somatic), H (flagellar), and K (capsular) antigens were the basis for the classification of EPEC strains. In 1987, when World Health Organization recommended that O26, O55, O86, O111, O114, O119, O125, O126, O127, O128, O142, and O158 should be considered as EPEC serogroups, some studies already reported that some of these serogroups included strains from other serotypes [14][25]. Serogroups such as O39, O88, O103, O145, O157, and O158 are now considered to be included in EPEC pathotype [26]. Among EPEC isolates, H2 and H6 are the most common flagellar antigens associated, but there are also some less common H types (such as H7, H8, H9, H12, H21, H27, H25, and H34), and some EPEC strains are classified as H negative (non-motile) [11][14][25][24][27]. Because of the high diversity of these antigens, serotyping is no longer considered a rapid diagnostic tool, nor a reliable one [13][28][29].
In 1982, EPEC was defined as “diarrheagenic E. coli belonging to serogroups epidemiologically incriminated as pathogens but whose pathogenic mechanisms have not been proven to be related either to heat-labile enterotoxins or heat-stable enterotoxins or to Shigella-like invasiveness” [30]. Now, EPEC strains are defined, based on their virulence factors, as diarrheagenic strains of E. coli that can produce attaching and effacing (A/E) lesions on the intestinal epithelium, but unable to produce Shiga toxins and heat-labile (LT) or heat-stable (ST) enterotoxins [3][11][24].
The most important characteristic of EPEC is the ability to produce A/E lesions, which enable them to cause localized lesions by attaching tightly to the surface of the intestinal epithelial cells, disrupting the cell surfaces, finally leading to the effacement of the microvilli. Intimin is an outer membrane protein, encoded by the eae gene, which mediates the intestinal cell attachment. The attachment is facilitated by Tir (Translocated intimin receptor), an effector inserted in the host plasma membrane, where it functions as a receptor for intimin. By a type-three-secretion-system (T3SS), EPEC can inject a large number (at least twenty-five) effector proteins in the host cells. All the required genetic elements for the A/E lesion are encoded on the locus of enterocyte effacement (LEE), a large genomic pathogenicity island [31][32][33][34].
EPEC can also be classified on typical and atypical isolates, based on the presence or absence of EAF (plasmid E. coli adherence factor), where bfp and per, two important operons, are localized. bfp encodes the type IV bundle-forming pilus (BFP), while per encodes the plasmid-encoded regulator (Per), a transcriptional activator [31]. All the EPEC strains are eae positive (+), so typical EPEC are described as eae+, bfpA+, while atypical strains are considered those eae+, bfpA− (negative) [11][27][31][35]. Some of the atypical EPEC strains might possess other virulence factors such as enteroaggregative heat-stable toxin (EAST1), hemolysin, not encoded on LEE [26].
3. Virulence Factors
EPEC can adhere, at least in vitro, to cell lines and organ cultures in a three-dimensional microcolonies pattern, named localized adherence (LA) pattern. BFP mediates LA phenotype, and it is also involved in antigenicity, biofilm formation, and autoaggregation [11]. EPEC are non-invasive pathogens that can induce lesions by introducing effector proteins directly into the host cells, using a type-three-secretion-system (T3SS). These effectors are encoded on the LEE pathogenicity island. LEE contains genes that encode T3SS (Esc, Sep), the outer membrane adhesin (intimin, essential for the adherence on the host cells, encoded by eae gene), translocators (EspA/B/D), effector proteins (EspF/G/H, Map, EspZ), chaperones (Ces proteins), the translocated intimin receptor (Tir), and regulatory proteins such as LEE-encoded regulator (Ler), a global regulator of LEE proteins (GrlA—activator, GrlR—repressor) [11][36][37][38][39][40].
Various non-LEE (Nle) encodes effector genes (cif, espI/nleA, nleB, nleC, nleD, nleE, nleH), located outside of the LEE region, clustered in six pathogenicity islands. The Nle proteins can increase the bacterial virulence by preventing/modulating the host inflammatory response and by disrupting the tight junctions and the cytoskeleton of the host cells [11][31][36][33]. Along with Nle A, EspF, and Map, other non-LEE encoded effector proteins are involved in the disassembly of the tight junctions [41].
In addition to BFP, some EPEC strains may present other fimbriae or pili (type 1 fimbriae, E. coli common pilus), flagella, astA gene (encoding EAST1—the enteroaggregative E. coli heat-stable enterotoxin 1), autotransporter proteins (EspC), or produce a hybrid adherence phenotype in HeLa cells (LA and aggregative-like pattern), but their involvement in the pathogenicity still needs to be studied [11][42].
4. Pathogenesis
Attaching-and-effacing was described by Nataro and Kaper, in 1998, as the “hallmark” of the EPEC infections, a lesion that is characterized by the intimate attachment of the bacteria to the host epithelial cell membrane and the effacement of the microvilli [25]. Since then, a lot of studies have focused on describing this main pathogenic mechanism, which is common to the typical and atypical EPEC strains. The interaction between EPEC and the host cells is described as a four-stage process (Figure 1).
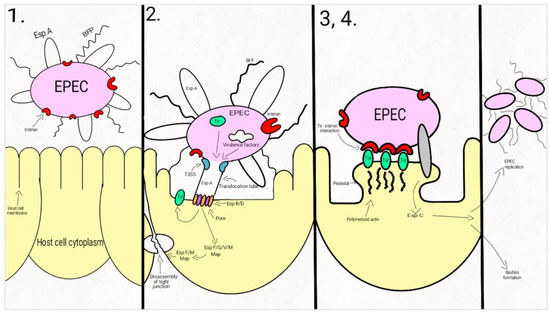
Figure 1. Schematic representation of the EPEC adherence mechanism: 1. EPEC express Bfp and EspA; 2. After attaching to the enterocyte (trough Bfp), EPEC use a T3SS to infect many effectors in the cell. The phosphorylated Tir is inserted into the host cell membrane; 3. The bacterial intimin binds to the modified Tir, attaching the bacteria to the host cell. Actin and cytoskeletal elements are accumulated near the site of the bacterial adherence. EspC is inserted into the cell trough an autotransporter system, T5SS. 4. The cytoskeletal elements, accumulated near the site of the attachment, leads to the formation of the pedestal structure, characteristic for EPEC. In the first step, EPEC cells express the intimate adhesin intimin, Bfp (bundle-forming pili), and EspA (short filaments surface-associated). Environmental factors regulate the expression of these virulence factors, influencing the site of the bacterial colonization (small/large bowel) [43][44][45].
In the second step, EPEC strains adhere to the intestinal epithelium through Bfp and EspA, forming dense microcolonies on the cell’s surface, in a pattern described as localized adherence. The type-three-secretion-system creates a pore, enabling the bacteria to inject Tir and a large number (at least 25, up to 50) of effector molecules into the host cell. These effectors facilitate bacterial colonization, immune evasion, and regulate inflammatory response and host cell death. They also activate the host cell-signaling pathways, causing the alteration of the cytoskeleton, leading to the loss of the microvilli. After tyrosine-protein kinase and protein kinase A modifies Tir, it is inserted into the host cell membrane [43][44][45][46][47][48][49][50][51][52][53][54].
Enterocyte effacement and intimate bacterial attachment to the host cell characterizes the third step of EPEC infection. The bacterial cells lose the EspA filaments from their surface. The bacterial intimin (encoded by eae) binds to the modified Tir, causing the intimate attachment of the bacteria to the host cell. In this phase, actin and cytoskeletal elements are accumulated near the site of the bacterial adherence [44][43][55]. An autotransporter system, T5SS (type V secretion system), mediates the secretion of EspC, [42][56][57][58], a protein involved not only in epithelial cell cytotoxicity, but also in bacterial replication and biofilm formation [56][59][60][61].
During the fourth step, the cytoskeletal elements, accumulated near the site of the attachment, leads to the formation of the pedestal structure, characteristic for EPEC. The effector molecules (translocated from the bacteria) disrupt the cell processes, leading, eventually, to cell death [24][43][62].
The clinical symptoms of EPEC diarrheal disease occurs before the complete establishment of the A/E lesions and the loss of the microvilli, so these mechanisms are most likely involved in the exacerbation of the diarrhea. The rapid onset of the diarrhea (sometimes only after a few hours after the ingestion) is more likely to be the result of a secretory mechanism [63][64].
EPEC can imbalance the host cells electrolyte transport, and by the dysregulation of these transport pathways, it is responsible for the rapid onset of the symptoms. Na+/H+ (NHE2-3), Na+/glucose (SGLT-1), and Cl−/HCO3− (DRA/PAT1) exchangers are responsible for the water and solutes intestinal absorption. CFTR (the apical cAMP-dependent cystic fibrosis transmembrane conductance regulator) is responsible, among other factors, for the osmotic gradient which controls the movement of water into the intestinal lumen. EPEC establish a disequilibrium in the Na+/Cl− electroneutral exchange through the plasma membrane, reducing water absorption. Moreover, by modulating the AQP (epithelial aquaporin) expression, the water transport is directly altered by EPEC [63][65][66][67][68].
EPEC is capable of disrupting the epithelial barrier function and structure. By activating MLC kinase, EspB-mediated phosphorylation, activation of PKCα, EPEC can increase the paracellular permeability. EPEC is also responsible for the disruption of epithelial apical junctional complexes and the alteration of the tight junctional proteins [63].
The inflammatory response is considered more likely to be involved in the severity and the duration of the disease than in the early stages of the infection. An increase of IL-1β, TNFα, interferon (IFN) γ in the infected mucosa is associated with the EPEC infection. Both pro-inflammatory and anti-inflammatory pathways of the epithelial cells are involved in the EPEC mediated inflammatory response. The EPEC secreted components are responsible for the pro-inflammatory response, while the effectors inserted in the host cells by the T3SS attenuates the inflammatory response [49][63][69].
References
- Troeger, C.; Blacker, B.F.; Khalil, I.A.; Rao, P.C.; Cao, S.; Zimsen, S.R.; Albertson, S.B.; Stanaway, J.D.; Deshpande, A.; Abebe, Z.; et al. Estimates of the Global, Regional, and National Morbidity, Mortality, and Aetiologies of Diarrhoea in 195 Countries: A Systematic Analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018, 18, 1211–1228, doi:10.1016/S1473-3099(18)30362-1.
- Yu, J.; Jing, H.; Lai, S.; Xu, W.; Li, M.; Wu, J.; Liu, W.; Yuan, Z.; Chen, Y.; Zhao, S.; et al. Etiology of Diarrhea among Children under the Age Five in China: Results from a Five-Year Surveillance. J. Infect. 2015, 71, 19–27, doi:10.1016/j.jinf.2015.03.001.
- Hazen, T.H.; Donnenberg, M.S.; Panchalingam, S.; Antonio, M.; Hossain, A.; Mandomando, I.; Ochieng, J.B.; Ramamurthy, T.; Tamboura, B.; Qureshi, S.; et al. Genomic Diversity of EPEC Associated with Clinical Presentations of Differing Severity. Nat. Microbiol. 2016, 1, 15014, doi:10.1038/nmicrobiol.2015.14.
- Radlović, N.; Leković, Z.; Vuletić, B.; Radlović, V.; Simić, D. Acute Diarrhea in Children. Srp. Arh. Celok. Lek. 2015, 143, 755–762, doi:10.2298/sarh1512755r.
- Bellido-Blasco, J.B.; Arnedo-Pena, A. Epidemiology of Infectious Diarrhea. Encycl. Environ. Health 2011, 659–671, doi:10.1016/B978-0-444-63951-6.00689-6.
- Allocati, N.; Masulli, M.; Alexeyev, M.F.; Di Ilio, C. Escherichia Coli in Europe: An Overview. Int. J. Environ. Res. Public Health 2013, 10, 6235–6254, doi:10.3390/ijerph10126235.
- Kotloff, K.L. The Burden and Etiology of Diarrheal Illness in Developing Countries. Pediatric Clin. North Am. 2017, 64, 799–814, doi:10.1016/j.pcl.2017.03.006.
- Robins-Browne, R.M.; Hartland, E.L. Escherichia Coli as a Cause of Diarrhea. J. Gastroenterol. Hepatol. 2002, 17, 467–475, doi:10.1046/j.1440-1746.2002.02769.x.
- Escherich, T. The Intestinal Bacteria of the Neonate and Breast-Fed Infant. 1884. Rev. Infect. Dis. 1988, 10, 1220–1225, doi:10.1093/clinids/10.6.1220.
- Chaudhuri, R.R.; Henderson, I.R. The Evolution of the Escherichia Coli Phylogeny. Infect. Genet. Evol. 2012, 12, 214–226, doi:10.1016/j.meegid.2012.01.005.
- Gomes, T.A.T.; Elias, W.P.; Scaletsky, I.C.A.; Guth, B.E.C.; Rodrigues, J.F.; Piazza, R.M.F.; Ferreira, L.C.S.; Martinez, M.B. Diarrheagenic Escherichia Coli. Braz. J. Microbiol. 2016, 47, 3–30, doi:10.1016/j.bjm.2016.10.015.
- Fratamico, P.M.; DebRoy, C.; Liu, Y.; Needleman, D.S.; Baranzoni, G.M.; Feng, P. Advances in Molecular Serotyping and Subtyping of Escherichia Coli. Front. Microbiol. 2016, 7, doi:10.3389/fmicb.2016.00644.
- DebRoy, C.; Fratamico, P.M.; Roberts, E. Molecular Serogrouping of Escherichia Coli. Anim. Health Res. Rev. 2018, 1–16, doi:10.1017/S1466252317000093.
- Campos, L.C.; Franzolin, M.R.; Trabulsi, L.R. Diarrheagenic Escherichia Coli Categories among the Traditional Enteropathogenic E. Coli O Serogroups: A Review. Memórias Do Inst. Oswaldo Cruz 2004, 99, 545–552, doi:10.1590/S0074-02762004000600001.
- Díaz-Jiménez, D.; García-Meniño, I.; Herrera, A.; García, V.; López-Beceiro, A.M.; Alonso, M.P.; Blanco, J.; Mora, A. Genomic Characterization of Escherichia Coli Isolates Belonging to a New Hybrid AEPEC/ExPEC Pathotype O153:H10-A-ST10 Eae-Beta1 Occurred in Meat, Poultry, Wildlife and Human Diarrheagenic Samples. Antibiotics 2020, 9, doi:10.3390/antibiotics9040192.
- Abdalhamid, B.; Mccutchen, E.L.; Bouska, A.C.; Weiwei, Z.; Loeck, B.; Hinrichs, S.H.; Iwen, P.C. Whole Genome Sequencing to Characterize Shiga Toxin-Producing Escherichia Coli O26 in a Public Health Setting. J. Infect. Public Health 2019, 12, 884–889, doi:10.1016/j.jiph.2019.06.008.
- Robins-Browne, R.M.; Holt, K.E.; Ingle, D.J.; Hocking, D.M.; Yang, J.; Tauschek, M. Are Escherichia Coli Pathotypes Still Relevant in the Era of Whole-Genome Sequencing? Front. Cell Infect. Microbiol. 2016, 6, doi:10.3389/fcimb.2016.00141.
- Koutsoumanis, K.; Allende, A.; Alvarez‐Ordóñez, A.; Bolton, D.; Bover‐Cid, S.; Chemaly, M.; Davies, R.; Cesare, A.D.; Hilbert, F.; Lindqvist, R.; et al. Whole Genome Sequencing and Metagenomics for Outbreak Investigation, Source Attribution and Risk Assessment of Food-Borne Microorganisms. Efsa J. 2019, 17, e05898, doi.10.2903/j.efsa.2019.5898.
- Arimizu, Y.; Kirino, Y.; Sato, M.P.; Uno, K.; Sato, T.; Gotoh, Y.; Auvray, F.; Brugere, H.; Oswald, E.; Mainil, J.G.; et al. Large-Scale Genome Analysis of Bovine Commensal Escherichia Coli Reveals That Bovine-Adapted E. Coli Lineages Are Serving as Evolutionary Sources of the Emergence of Human Intestinal Pathogenic Strains. Genome Res. 2019, 29, 1495–1505, doi:10.1101/gr.249268.119.
- Iguchi, A.; Thomson, N.R.; Ogura, Y.; Saunders, D.; Ooka, T.; Henderson, I.R.; Harris, D.; Asadulghani, M.; Kurokawa, K.; Dean, P.; et al. Complete Genome Sequence and Comparative Genome Analysis of Enteropathogenic Escherichia Coli O127:H6 Strain E2348/69. J. Bacteriol. 2009, 191, 347–354, doi:10.1128/JB.01238-08.
- Kaper, J.B. Pathogenic Escherichia Coli. Int. J. Med. Microbiol. 2005, 295, 355–356, doi:10.1016/j.ijmm.2005.06.008.
- Chervy, M.; Barnich, N.; Denizot, J. Adherent-Invasive E. Coli: Update on the Lifestyle of a Troublemaker in Crohn’s Disease. Int. J. Mol. Sci. 2020, 21, doi:10.3390/ijms21103734.
- Meza-Segura, M.; Estrada-Garcia, T. Diffusely Adherent Escherichia Coli. In Escherichia Coli in the Americas; Torres, A.G., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 125–147. ISBN 978-3-319-45092-6.
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent Advances in Understanding Enteric Pathogenic Escherichia Coli. Clin. Microbiol. Rev. 2013, 26, 822–880, doi:10.1128/CMR.00022-13.
- Nataro, J.P.; Kaper, J.B. Diarrheagenic Escherichia Coli. Clin. Microbiol. Rev. 1998, 11, 142–201, doi:10.1128/CMR.11.1.142.
- Hu, J.; Torres, A.G. Enteropathogenic Escherichia Coli: Foe or Innocent Bystander? Clin Microbiol. Infect. 2015, 21, 729–734, doi:10.1016/j.cmi.2015.01.015.
- Trabulsi, L.R.; Keller, R.; Gomes, T.A.T. Typical and Atypical Enteropathogenic Escherichia Coli. Emerg. Infect. Dis. J. Cdc 2002, 8, doi:10.3201/eid0805.010385.
- Sánchez, S.; Llorente, M.T.; Echeita, M.A.; Herrera-León, S. Development of Three Multiplex PCR Assays Targeting the 21 Most Clinically Relevant Serogroups Associated with Shiga Toxin-Producing E. Coli Infection in Humans. PLoS ONE 2015, 10, doi:10.1371/journal.pone.0117660.
- Joensen, K.G.; Tetzschner, A.M.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and Easy In Silico Serotyping of Escherichia Coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 2410–2426, doi:10.1128/JCM.00008-15.
- Scaletsky, I.C.; Silva, M.L.; Toledo, M.R.; Davis, B.R.; Blake, P.A.; Trabulsi, L.R. Correlation between Adherence to HeLa Cells and Serogroups, Serotypes, and Bioserotypes of Escherichia Coli. Infect. Immun. 1985, 49, 528–532.
- Ochoa, T.J.; Contreras, C.A. Enteropathogenic E. Coli (EPEC) Infection in Children. Curr. Opin. Infect. Dis. 2011, 24, 478.
- Singh, A.P.; Aijaz, S. Enteropathogenic E. Coli: Breaking the Intestinal Tight Junction Barrier. F1000Res 2016, 4, 231, doi:10.12688/f1000research.6778.2.
- Cepeda-Molero, M.; Berger, C.N.; Walsham, A.D.S.; Ellis, S.J.; Wemyss-Holden, S.; Schüller, S.; Frankel, G.; Fernández, L.Á. Attaching and Effacing (A/E) Lesion Formation by Enteropathogenic E. Coli on Human Intestinal Mucosa Is Dependent on Non-LEE Effectors. PLoS Pathog. 2017, 13, e1006706, doi:10.1371/journal.ppat.1006706.
- de Jong, M.F.; Alto, N.M. Cooperative Immune Suppression by Escherichia Coli and Shigella Effector Proteins. Infect. Immun. 2018, 86, doi:10.1128/IAI.00560-17.
- Montso, P.K.; Mlambo, V.; Ateba, C.N. The First Isolation and Molecular Characterization of Shiga Toxin-Producing Virulent Multi-Drug Resistant Atypical Enteropathogenic Escherichia Coli O177 Serogroup From South African Cattle. Front. Cell. Infect. Microbiol. 2019, 9, doi:10.3389/fcimb.2019.00333.
- Dean, P.; Kenny, B. The Effector Repertoire of Enteropathogenic E. Coli: Ganging up on the Host Cell. Curr. Opin. Microbiol. 2009, 12, 101–109, doi:10.1016/j.mib.2008.11.006.
- Litvak, Y.; Sharon, S.; Hyams, M.; Zhang, L.; Kobi, S.; Katsowich, N.; Dishon, S.; Nussbaum, G.; Dong, N.; Shao, F.; et al. Epithelial Cells Detect Functional Type III Secretion System of Enteropathogenic Escherichia Coli through a Novel NF-ΚB Signaling Pathway. PLoS Pathog. 2017, 13, e1006472, doi:10.1371/journal.ppat.1006472.
- Pinaud, L.; Sansonetti, P.J.; Phalipon, A. Host Cell Targeting by Enteropathogenic Bacteria T3SS Effectors. Trends Microbiol. 2018, 26, 266–283, doi:10.1016/j.tim.2018.01.010.
- Leh, H.; Khodr, A.; Bouger, M.-C.; Sclavi, B.; Rimsky, S.; Bury-Moné, S. Bacterial-Chromatin Structural Proteins Regulate the Bimodal Expression of the Locus of Enterocyte Effacement (LEE) Pathogenicity Island in Enteropathogenic Escherichia Coli. mBio 2017, 8, doi:10.1128/mBio.00773-17.
- Pal, R.R.; Baidya, A.K.; Mamou, G.; Bhattacharya, S.; Socol, Y.; Kobi, S.; Katsowich, N.; Ben-Yehuda, S.; Rosenshine, I. Pathogenic E. Coli Extracts Nutrients from Infected Host Cells Utilizing Injectisome Components. Cell 2019, 177, 683–696.e18, doi:10.1016/j.cell.2019.02.022.
- Navarro-Garcia, F.; Serapio-Palacios, A.; Ugalde-Silva, P.; Tapia-Pastrana, G.; Chavez-Dueñas, L. Actin Cytoskeleton Manipulation by Effector Proteins Secreted by Diarrheagenic Escherichia Coli Pathotypes. Available online: https://www.hindawi.com/journals/bmri/2013/374395/ (accessed on 27 December 2020).
- Guignot, J.; Segura, A.; Tran Van Nhieu, G. The Serine Protease EspC from Enteropathogenic Escherichia Coli Regulates Pore Formation and Cytotoxicity Mediated by the Type III Secretion System. PLoS Pathog. 2015, 11, e1005013, doi:10.1371/journal.ppat.1005013.
- Clarke, S.C.; Haigh, R.D.; Freestone, P.P.E.; Williams, P.H. Virulence of Enteropathogenic Escherichia Coli, a Global Pathogen. Clin. Microbiol. Rev. 2003, 16, 365–378, doi:10.1128/CMR.16.3.365-378.2003.
- Vallance, B.A.; Finlay, B.B. Exploitation of Host Cells by Enteropathogenic Escherichia Coli. PNAS 2000, 97, 8799–8806, doi:10.1073/pnas.97.16.8799.
- Franzin, F.M.; Sircili, M.P. Locus of Enterocyte Effacement: A Pathogenicity Island Involved in the Virulence of Enteropathogenic and Enterohemorragic Escherichia Coli Subjected to a Complex Network of Gene Regulation. Biomed. Res. Int. 2015, 2015, doi:10.1155/2015/534738.
- Donnenberg, M.S.; Kaper, J.B. Enteropathogenic Escherichia Coli. Infect. Immun. 1992, 60, 3953–3961.
- Scholz, R.; Imami, K.; Scott, N.E.; Trimble, W.S.; Foster, L.J.; Finlay, B.B. Novel Host Proteins and Signaling Pathways in Enteropathogenic E. Coli Pathogenesis Identified by Global Phosphoproteome Analysis. Mol. Cell. Proteom. 2015, 14, 1927–1945, doi:10.1074/mcp.M114.046847.
- Hartland, E.L.; Leong, J. Enteropathogenic and Enterohemorrhagic E. Coli: Ecology, Pathogenesis, and Evolution. Front. Cell. Infect. Microbiol. 2013, 3, doi:10.3389/fcimb.2013.00015.
- Shenoy, A.R.; Furniss, R.C.D.; Goddard, P.J.; Clements, A. Modulation of Host Cell Processes by T3SS Effectors. Curr. Top. Microbiol. Immunol. 2018, 416, 73–115, doi:10.1007/82_2018_106.
- Song, T.; Li, K.; Zhou, W.; Zhou, J.; Jin, Y.; Dai, H.; Xu, T.; Hu, M.; Ren, H.; Yue, J.; et al. A Type III Effector NleF from EHEC Inhibits Epithelial Inflammatory Cell Death by Targeting Caspase-4. Biomed. Res. Int. 2017, 2017, doi:10.1155/2017/4101745.
- Pearson, J.S.; Giogha, C.; Ong, S.Y.; Kennedy, C.L.; Kelly, M.; Robinson, K.S.; Wong, T.; Mansell, A.; Riedmaier, P.; Oates, C.V.; et al. A Type III Effector Antagonises Death Receptor Signalling during Bacterial Gut Infection. Nature 2013, 501, 247–251, doi:10.1038/nature12524.
- Wong, A.R.C.; Pearson, J.S.; Bright, M.D.; Munera, D.; Robinson, K.S.; Lee, S.F.; Frankel, G.; Hartland, E.L. Enteropathogenic and Enterohaemorrhagic Escherichia Coli: Even More Subversive Elements. Mol. Microbiol. 2011, 80, 1420–1438, doi:10.1111/j.1365-2958.2011.07661.x.
- Ugalde-Silva, P.; Gonzalez-Lugo, O.; Navarro-Garcia, F. Tight Junction Disruption Induced by Type 3 Secretion System Effectors Injected by Enteropathogenic and Enterohemorrhagic Escherichia Coli. Front. Cell Infect. Microbiol. 2016, 6, doi:10.3389/fcimb.2016.00087.
- Creuzburg, K.; Giogha, C.; Wong Fok Lung, T.; Scott, N.E.; Mühlen, S.; Hartland, E.L.; Pearson, J.S. The Type III Effector NleD from Enteropathogenic Escherichia Coli Differentiates between Host Substrates P38 and JNK. Infect. Immun. 2017, 85, doi:10.1128/IAI.00620-16.
- Deborah Chen, H.; Frankel, G. Enteropathogenic Escherichia Coli: Unravelling Pathogenesis. FEMS Microbiol. Rev. 2005, 29, 83–98, doi:10.1016/j.femsre.2004.07.002.
- Serapio-Palacios, A.; Navarro-Garcia, F. EspC, an Autotransporter Protein Secreted by Enteropathogenic Escherichia Coli, Causes Apoptosis and Necrosis through Caspase and Calpain Activation, Including Direct Procaspase-3 Cleavage. mBio 2016, 7, doi:10.1128/mBio.00479-16.
- Sanchez-Villamil, J.I.; Navarro-Garcia, F.; Castillo-Romero, A.; Gutierrez-Gutierrez, F.; Tapia, D.; Tapia-Pastrana, G. Curcumin Blocks Cytotoxicity of Enteroaggregative and Enteropathogenic Escherichia Coli by Blocking Pet and EspC Proteolytic Release From Bacterial Outer Membrane. Front. Cell Infect. Microbiol. 2019, 9, doi:10.3389/fcimb.2019.00334.
- Habouria, H.; Pokharel, P.; Maris, S.; Garénaux, A.; Bessaiah, H.; Houle, S.; Veyrier, F.J.; Guyomard-Rabenirina, S.; Talarmin, A.; Dozois, C.M. Three New Serine-Protease Autotransporters of Enterobacteriaceae (SPATEs) from Extra-Intestinal Pathogenic Escherichia Coli and Combined Role of SPATEs for Cytotoxicity and Colonization of the Mouse Kidney. Virulence 2019, 10, 568–587, doi:10.1080/21505594.2019.1624102.
- Navarro-Garcia, F.; Serapio-Palacios, A.; Vidal, J.E.; Salazar, M.I.; Tapia-Pastrana, G. EspC Promotes Epithelial Cell Detachment by Enteropathogenic Escherichia Coli via Sequential Cleavages of a Cytoskeletal Protein and Then Focal Adhesion Proteins. Infect. Immun. 2014, 82, 2255–2265, doi:10.1128/IAI.01386-13.
- Govindarajan, D.K.; Viswalingam, N.; Meganathan, Y.; Kandaswamy, K. Adherence Patterns of Escherichia Coli in the Intestine and Its Role in Pathogenesis. Med. Microecol. 2020, 5, 100025, doi:10.1016/j.medmic.2020.100025.
- Xicohtencatl-Cortes, J.; Saldaña, Z.; Deng, W.; Castañeda, E.; Freer, E.; Tarr, P.I.; Finlay, B.B.; Puente, J.L.; Girón, J.A. Bacterial Macroscopic Rope-like Fibers with Cytopathic and Adhesive Properties. J. Biol. Chem. 2010, 285, 32336–32342, doi:10.1074/jbc.M110.162248.
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia Coli. Nat. Rev. Microbiol. 2004, 2, 123–140, doi:10.1038/nrmicro818.
- Lapointe, T.K.; O’Connor, P.M.; Buret, A.G. The Role of Epithelial Malfunction in the Pathogenesis of Enteropathogenic E. Coli -Induced Diarrhea. Lab. Investig. 2009, 89, 964–970, doi:10.1038/labinvest.2009.69.
- Gujral, T.; Kumar, A.; Priyamvada, S.; Saksena, S.; Gill, R.K.; Hodges, K.; Alrefai, W.A.; Hecht, G.A.; Dudeja, P.K. Mechanisms of DRA Recycling in Intestinal Epithelial Cells: Effect of Enteropathogenic E. Coli. Am. J. Physiol. Cell Physiol. 2015, 309, C835–C846, doi:10.1152/ajpcell.00107.2015.
- Cao, L.; Yuan, Z.; Liu, M.; Stock, C. (Patho-)Physiology of Na+/H+ Exchangers (NHEs) in the Digestive System. Front. Physiol. 2020, 10, doi:10.3389/fphys.2019.01566.
- Gurney, M.A.; Laubitz, D.; Ghishan, F.K.; Kiela, P.R. Pathophysiology of Intestinal Na+/H+ Exchange. Cell Mol. Gastroenterol. Hepatol. 2016, 3, 27–40, doi:10.1016/j.jcmgh.2016.09.010.
- Das, S.; Jayaratne, R.; Barrett, K.E. The Role of Ion Transporters in the Pathophysiology of Infectious Diarrhea. Cell Mol. Gastroenterol. Hepatol. 2018, 6, 33–45, doi:10.1016/j.jcmgh.2018.02.009.
- Anand, S.; Mandal, S.; Patil, P.; Tomar, S.K. Pathogen-Induced Secretory Diarrhea and Its Prevention. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1721–1739, doi:10.1007/s10096-016-2726-5.
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of Assembly, Regulation and Signalling. Nat. Rev. Immunol. 2016, 16, 407–420, doi:10.1038/nri.2016.58.




