
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jianbin Xiong | + 2713 word(s) | 2713 | 2021-01-25 11:12:59 | | | |
| 2 | Catherine Yang | -3 word(s) | 2710 | 2021-03-04 03:31:21 | | | | |
| 3 | Lily Guo | + 2 word(s) | 2712 | 2021-03-04 04:57:26 | | |
Video Upload Options
Plant phenotypic image recognition (PPIR) is an important branch of smart agriculture. In recent years, deep learning has achieved significant breakthroughs in image recognition.
1. Introduction
Plants are indispensable resources that are present on the earth. They play an important role in the development of the society and they have great significance in environmental protection, medical pharmaceutical, agricultural development, and food-related applications [1]. However, any plant-related work, such as plant species and diseases identification and evaluation of plant production, is becoming increasingly complex. An important starting point for any plant-related work is the identification of plant phenotype that refers to the physiological and biochemical characteristics of plants, including their color, shape, texture, and so on, which are determined by both genes and the environment. Traditional methods of plant phenotype identification include artificial identification, phytochemical classification, the anatomical method, morphological method, and genetic method, which are difficult to implement, have low efficiency, and unstable accuracy [2]. With the development and popularity of computer technology, image recognition technology is becoming increasingly mature, and it has been successfully applied in many fields, such as face recognition, object detection, medical imaging, etc. [3][4]. Plant phenotype identification tht is based on image processing technology has become a popular topic of research, leading to new breakthroughs and improved accuracy. In particular, deep learning has been proposed in order to further promote the development of PPIR[5]. Table 1 shows recent relevant reviews.
| References | Review Main Points |
|---|---|
| Muhammad et al. [6] | This paper aims to review and analyze the implementation and performance of various methodologies (artificial neural network (ANN), probabilistic neural network (PNN), convolutional neural network (CNN), K-nearest neighbor (KNN) and support vector machine (SVM)) on plant classification. At the same time including feature extraction and preprocessing technology. Each technique has its advantages and limitations in leaf pattern recognition. The quality of leaf images plays an important role, and therefore, a reliable source of leaf database must be used to establish the machine learning algorithm prior to leaf recognition and validation. |
| Weng et al.[7] | In this survey, authors elaborate the wor k from four different aspects: (1) plant morphology and physiological information extraction, (2) plant identification and weed detection, (3) pest detection, and (4) yield prediction. It focuses on the specific application of convolutional neural networks in this field. Authors also analyze the pros and cons of these methods compared to traditional approaches. The potential future trends of plant phenotyping research are discussed at the end of this survey. |
| Wang et al.[1] | The review introduces the research significance and history of plant recognition technologies. Then, the main technologies and steps of plant recognition are reviewed. At the same time, more than 30 leaf features (including 16 shape features, 11 texture features, four color features), and then SVM was used to evaluate these features and their fusion features, and 8 commonly used classifiers are introduced in detail. Finally, the review is ended with a conclusion of the insufficient of plant identification technologies and a prediction of future development. |
| Barbedo [8] | This paper provides an analysis of each one of those challenges, emphasizing both the problems that they may cause and how they may have potentially affected the techniques proposed in the past. Some possible solutions capable of overcoming at least some of those challenges are proposed. Focusing on plant diseases, automatic identification, visible symptoms, digital image processing, extrinsic factors (image background, image capture conditions), intrinsic factors (symptom segmentation, symptom variations, multiple simultaneous disorders, different disorders with similar symptoms), other challenges and future prospects. |
| Cope et al. [9] | The authors review the main computational, morphometric and image processing methods that have been used in recent years to analyze images of plants, introducing readers to relevant botanical concepts along the way. They discuss the measurement of leaf outlines, flower shape, vein structures and leaf textures, and describe a wide range of analytical methods in use. At last, they discuss a number of systems that apply this research, including prototypes of hand-held digital field guides and various robotic systems used in agriculture. They conclude with a discussion of ongoing work and outstanding problems in the area. |
| Waldchen et al.[10] | This paper is the first systematic literature review with the aim of a thorough analysis and comparison of primary studies on computer vision approaches for plant species identification. They identified 120 peer-reviewed studies, selected through a multi-stage process, published in the last 10 years (2005–2015). After a careful analysis of these studies, they describe the applied methods categorized according to the studied plant organ, and the studied features, i.e., shape, texture, color, margin, and vein structure. Furthermore, they compare methods based on classification accuracy achieved on publicly available datasets. Their results are relevant to researches in ecology as well as computer vision for their ongoing research. |
| Thyagharajan et al. [11] | Authors review several image processing methods in the feature extraction of leaves, given that feature extraction is a crucial technique in computer vision. As computers cannot comprehend images, they are required to be converted into features by individually analyzing image shapes, colors, textures and moments. Images that look the same may deviate in terms of geometric and photometric variations. In their study, they also discuss certain machine learning classifiers for an analysis of different species of leaves. |
| This paper | In this paper, three categories of plant image recognition algorithms are summarized, and the methods of plant image preprocessing and plant image feature extraction are summarized. Then, the advantages and disadvantages of imaging technologies are explained. At last, the specific applications of four common deep learning models in plant image recognition are described. |
2. PPIR Technology
2.1. State of the Art in PPIR Technology
The development of PPIR technology started several decades earlier internationally, focusing on feature extraction and training of plants using traditional methods. In the 1980s and 1990s, Ingrouille et al. [12], from the University of London, extracted 27 main characteristics of plant leaves and used principal components analysis (PCA) in order to classify oak trees. Yonekawa et al.[13], from the University of Tokyo, fused several prominent features of plant phenotypes, such as texture, color, and shape for image recognition, and used the backpropagation (BP) neural network algorithm to train and classify image data. In 2006, Cheng et al. [14] used fuzzy functions for shape matching and identification of plant phenotypes. CLEF 2011–2015 (Cross Language Evaluation Forum) in the BBS held pictures of plant classification of image recognition under acomplex environment; the library has 1000 kinds of plant species. Villena et al.[15] utilized scale invariance to extract plant phenotypic traits that can be identified. In 2013, Charles et al. [16] established a database of 100 plants containing 16 samples for each plant, carried out feature extraction, and proposed a high-accuracy recognition algorithm under the condition of small training set size, based on the k-nearest neighbor (KNN) algorithm. When the shapes, textures, and edges of the plant phenotypes were fused, an accuracy of 96% was achieved.The research on PPIR started late domestically, but it is worth learning from. In 2007, Wang et al. [17]used a moving center hypersphere classifier to classify eight geometric features and seven image invariant moments that were extracted from ginkgo leaves with an accuracy rate of 92%. In 2009, Wang et al.[18] extracted the feature vectors of maize leaves while using morphology and contour extraction, and then classified them while using the genetic algorithm for optimized selection of the features. Subsequently, Fisher’s discrimination method was used in order to identify the diseased leaves with an accuracy rate of more than 90%. In Reference [19], Zhai et al. used the relational matching structure method to match the plant leaves images and different model structures after feature extraction, and identified the types of plants based on the matching level. In 2015, Wang et al.[20] proposed a plant leaves fusion-based recognition system to extract the development characteristics of a variety of foliage plant phenotypic traits, such as shape, color, texture, leaf margin, etc. Support vector machine (SVM) classification was used for plant identification, and the experimental results showed that an accuracy of 91.41% was achieved while using the SVM, which was better than that with a BP neural network or the KNN algorithm. In spectroscopy, Cen et al.[21] used hyperspectral imaging technology in combination with supervised classification algorithm for cucumber freezing damage detectio2.2. Traditional PPIR TechniquesThe existing PPIR methods can be mainly divided into three categories[3], which are described, as follows:(1) the basic idea of relational structure matching method for PPIR is shown in Figure 1[22]. In this method, first, the input images are preprocessed in order to extract features, while using multi-scale curvature space to describe the geometric features, as well as the fuzzy particle swarm algorithm and genetic algorithm. Second, the algorithm matching rules and parameters are set. Finally, the extracted features are matched with the features from the sample database and images are classified based on the matching degree[23][24].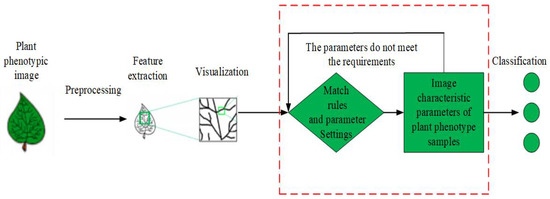 Figure 1. Flow chart of relational structure matching method.
Figure 1. Flow chart of relational structure matching method.
(2) PPIR that is based on mathematical statistics is the most widely used method. Figure 2 shows its basic idea. First, a mathematical model is set up, followed by quantitative analysis and classification of the image. The methods in this category are based on Bayesian discriminant functions, KNN, kernel PCA, Fisher discriminant method, etc. [25][26][27].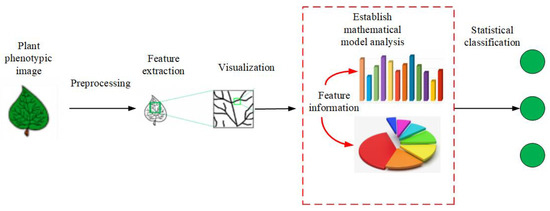 Figure 2. The flow chart of mathematical statistics methods.
Figure 2. The flow chart of mathematical statistics methods.
(3) Traditional machine learning-based PPIR mainly consists of artificial neural network, moving center hypersphere classifier, SVM, etc.[28]. Machine learning refers to a set of computerized modeling methods whose patterns are learned from data in order to automatically make decisions without explicit rules. The main idea of machine learning is to make effective use of experience or sample scenarios to discover the underlying structure, similarity, or difference in the data, so as to correctly interpret or classify new experience or sample scenarios[29]. It is important for programers to deploy specific machine learning approaches to their specific problems to make informed choices. The application of plant phenotype can be summarized into four aspects: (a) identification and detection, (b) classification, (c) quantification and estimation, and (d) prediction. In addition, data preprocessing steps, such as dimensionality reduction, clustering, and segmentation, can also be the key to a successful decision [29]. The moving center hypersphere classifier considers the sample points of plant phenotypic image data as a series of hyper spheres. A set of sample points are considered to be part of a hyper sphere, whose radius is expanded to include as many sample points as possible[26]. The SVM is a supervised learning model that is applicable to linearly or nonlinearly separable and a small number of samples. The method can be extended to high-dimensional pattern recognition by projecting the data points into a higher dimensional space and computing a maximum-margin hyperplane decision surface[26]. The SVM can be used to classify the plant phenotypic image data. Figure 3 shows its basic idea.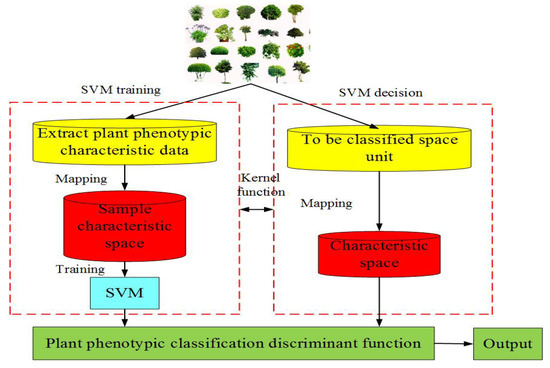 Figure 3. Flow chart of support vector machine (SVM).
Figure 3. Flow chart of support vector machine (SVM).
Feature extraction involving the extraction of shape, texture, color, and other major feature information is an important step in PPIR [22]. In shape based feature learning, edge detection and shape context description methods are widely used in order to extract the plant contours from the input images to achieve plant recognition [19]. Texture-based feature learning includes internal information of plant phenotypes and, generally, it is based on a local binary pattern (LBP) algorithm that calculates the correlation between a pixel and its surrounding pixels in an object [24]. Color based feature learning is more stable and reliable when compared with the aforementioned learning methods. It is robust and not sensitive to the target size and orientation of the color characteristics. It usually uses the percentage of pixels of different colors in red, green, and blue (RGB), or hue, saturation, and brightness (HSV) images, and their histograms for feature extraction and image recognition [25]. These feature learning methods focus on the attributes of plant phenotypes and they mostly include shallow learning methods that need manual feature extraction.At present, a variety of imaging technologies are used in order to collect complex traits that are related to growth, yield, and adaptability of biotic or abiotic stresses (such as disease, insects, drought, and salinity). These imaging technologies include visible light imaging (such as machine vision), imaging spectroscopy (such as multispectral and hyperspectral remote sensing), thermal infrared imaging, fluorescence imaging, 3D imaging and tomography (such as positron emission computer tomography), and image and computer tomography). Many institutions and organizations in the world have carried out phenotypic group analysis, such as the Australian plant phenomics facility. At the same time, there are also some high-throughput phenotypic testing platforms that are deployed in the field or indoors, such as LemnaTec. Although phenotype analysis of plants that is based on optical imaging has many advantages, it also faces some difficulties. For example, when machine vision methods are used to process visible light images in order to obtain phenotypic information, such as plant species, fruit quantity, and pest categories, it is difficult to resolve adjacent leaves problems, such as overlap and occlusion that are caused by ears and fruits. Images that were collected in a laboratory environment often have a pure background, uniform lighting, and a small number of plants or organs contained in the image. Solving practical problems in the field is often caused by complex backgrounds, differences in lighting, and occlusion. The interference of object shadows.For PPIR, especially for a large database of plant phenotypic images, the performance of shallow and single feature learning methods is not satisfactory due to the low recognition accuracy and several interference factors [26].In Table 1, the advantages and disadvantages of traditional methods that are used for plant phenotypes image recognition are compared.Table 1. Comparison of traditional plant phenotype recognition techniques.
| Methodsand Techniques | Introduction | Advantages | Disadvantages |
|---|---|---|---|
| K-NearestNeighbor (KNN) [22] | KNN algorithm is a basic classification and regression method. In the field of plant phenotype recognition and classification, it mainly undertakes the tasks of feature information retrieval, clustering, information filtering, and species recognition. | 1. Simple algorithm and mature theory. 2. Robust with regard to search space. 3. No training is required, confidence level can be obtained. | 1. High memory and computational cost at testing stage. 2. Sometimes sensitive to noise or nonlinear input. 3. lazy learning. |
| ProbabilisticNeural Network (PNN) [16] | The PNN algorithm is a neural network model based on statistical principles. It is a parallel algorithm developed based on the Bayesian minimum risk criterion. Unlike the traditional multi-layer forward network, the BP algorithm needs to be used to calculate the backward error propagation. It is a completely forward calculation process and is often used in the task of plant phenotypic image classification. | 1. Strong adaptability to noisy input and variable data. 2. Can have multiple outputs. | 1. Complexstructure. 2. Susceptible to overfitting. |
| SupportVector Machines (SVM) [5] | The SVM algorithm is an excellent data mining technology. Its goal is to find the optimal hyperplane to minimize the classifier error. It is widely used in statistical classification and regression analysis. It usually assumes the role of feature classifier in plant phenotype image recognition. | 1. Good generalization. 2. Sparsity of the solution and capacity control obtained by optimizing the margin. 3. Strong fault tolerance ability, relatively stable even with training sample deviation. | 1. Complex algorithm structure. 2. Slow training speed. |
| Decision Trees (DT) [27] | The DT algorithm is a tree-like decision diagram with additional probability results. It is a predictive model that intuitively uses statistical probability analysis to represent a mapping between object attributes and object values. In the field of plant phenotype classification and recognition, it often undertakes analysis the task of collecting statistics on plant phenotypic characteristics. | 1. Simple to use and easy to understand. 2. Pruning strategy eliminates a large number of weak correlations and irrelevant information to improve efficiency. 3. Fast prediction ability. | Sensitive to subtle changes in the attribute value. |
| ArtificialNeural Network (ANN) [28] | ANN algorithm is a kind of simulated biological neural network, which is a kind of pattern matching algorithm. It usually used to solve classification and regression problems. It also used in plant phenotype image recognition. | 1. Strong robustness and fault tolerance. 2. Complex nonlinear relations can be modeled using one or more hidden layers. | 1. Slow convergence speed and high complexity. 2. Possibility of local overfitting. |
| Random Forest (RF) [30] | In machine learning, RF is a classifier containing multiple decision trees, and its output category is determined by the mode of the category output by individual trees. It often undertakes species classification tasks in the field of plant phenotypes. | 1. The algorithm can handle very high dimensional data without feature selection. 2. Fast training speed, and easy to parallelize method. 3. The algorithm has strong anti-interference ability and strong anti-overfitting ability. | 1. When the algorithm solves regression problems, it does not perform as well as it does in classification. 2. The internal part of the model is relatively complicated, and it can only be tried between different parameters and random seeds. 3. For small data or low-dimensional, it may not produce a good classification. |
References
- Wang, Z.B.; Li, H.L.; Zhu, Y.; Xu, T.F. Review of plant identification based on image processing. Arch. Comput. Methods Eng. 2017, 24, 637–654.
- Mohanty, S.P.; Hughes, D.P.; Salathé, M.L. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2016, 7, 14–19.
- Zhang, N.; Liu, W.P. Plant leaf recognition technology based on image analysis. Appl. Res. Comput. 2011, 18, 7–13.
- Javed, A.; Nour, M.; Hasnat, K.; Essam, D.; Waqas, H.; Abdul, W. A review of intrusion detection systems using machine and deep learning in internet of things: Challenges, solutions and future directions. Electronics 2020, 9, 1177.
- Ma, Z.Y. Research and Test on Plant Leaves Recognition System Based on Deep Learning and Support Vector Machnie; Inner Mongolia Agricultural University: Hohhot, China, 2016.
- Muhammad, A.F.A.; Lee, S.C.; Fakhrul, R.R.; Farah, I.A.; Sharifah, R.W.A. Review on techniques for plant leaf classification and recognition. Computers 2019, 8, 77.
- Weng, Y.; Zeng, R.; Wu, C.M.; Wang, M.; Wang, X.J.; Liu, Y.J. A survey on deep-learning-based plant phenotype research in agriculture. Sci. Sin. 2019, 49, 698–716.
- Barbedo, J.G.A. A review on the main challenges in automatic plant disease identification based on visible range images. Biosyst. Eng. 2016, 144, 52–60.
- Cope, J.S.; Corney, D.; Clark, J.Y.; Remagnino, P.; Wilkin, P. Plant species identification using digital morphometrics: A review. Expert Syst. Appl. 2012, 39, 7562–7573.
- Waldchen, J.; Mader, P. Plant Species Identification Using Computer Vision Techniques: A Systematic Literature Review. Arch. Comput. Methods Eng. 2016, 25, 507–543.
- Thyagharajan, K.K.; Raji, I.K. A Review of Visual Descriptors and Classification Techniques Used in Leaf Species Identification. Arch. Comput. Methods Eng. 2019, 26, 933–960.
- Ingrouille, M.J.; Laird, S.M. A quantitative approach to oak variability in some north London woodlands. Comput. Appl. Res. 1986, 65, 25–46.
- Yonekawa, S.; Sakai, N.; Kitani, O. Identification of idealied leaf types using simple dimensionless shape factors by image analysis. Trans. ASAE 1993, 39, 1525–1533.
- Cheng, S.C.; Jhao, J.J.; Liou, B.H. PHD plant search system based on the characteristics of leaves using fuzzy function. New Trends Artif. Intell. 2007, 5, 834–844.
- Villena, R.J.; Lana, S.S.; Cristobal, J.C.G. Daedalus at image CLEF 2011 plant identification task: Using SIFT keypoints for object detection. In Proceedings of the CLEF 2011 Labs and Workshop, Amsterdam, The Netherlands, 19–22 September 2011.
- Charles, M.; James, C.; James, O. Plant leaf classification using probabilistic integration of shape, texture and margin features. Comput. Graph. Imaging 2013, 5, 45–54.
- Wang, X.F.; Huang, D.S.; Du, J.X.; Zhang, G.J. Feature extraction and recognition for leaf images. Comput. Eng. Appl. 2006, 3, 190–193.
- Wang, N.; Wang, K.R.; Xie, R.Z.; Lai, J.C.; Ming, B.; Li, S.K. Maize leaf disease identification based on fisher discrimination analysis. Sci. Agric. Sin. 2009, 42, 3836–3842.
- Zhai, C.M.; Du, J.X. A plant leaf image matching method based on shape context features. J. Guangxi Norm. Univ. (Nat. Sci. Ed.) 2009, 27, 171–174.
- Wang, L.J.; Huai, Y.J.; Peng, Y.C. Method of identification of foliage from plants based on extraction of multiple features of leaf images. J. Beijing For. Univ. 2015, 37, 55–61.
- Cen, H.Y.; Lu, R.F.; Zhu, Q.B.; Fernando, M. Nondestructive detection of chilling injury in cucumber fruit using hyperspectral imaging with feature selection and supervised classification. Postharvest Biol. Technol. 2016, 111, 352–361.
- Zhang, S. Research on Plant Leaf Images Identification Algorithm Based on Deep Learning; Beijing Forestry University: Beijing, China, 2016.
- Osikar, J.O. Computer Vision Classification of Leaves from Swedish Trees; Linkoing University: Linköping, Sweden, 2001.
- Iqbal, Z.; Khan, M.A.; Sharif, M.; Shah, J.H.; Rehman, M.H.; Javed, K. An automated detection and classification of citrus plant diseases using image processing techniques: A review. Comput. Electron. Agric. 2018, 152, 12–32.
- Zhao, Q.Y.; Kong, P.; Min, J.Z. A review of deep learning methods for the detection and classification of pulmonary nodules. J. Biomed. Eng. 2019, 36, 1060–1068.
- Huo, Y.Q.; Zhang, C.; Li, Y.H.; Zhi, W.T.; Zhang, J. Nondestructive detection for kiwifruit based on the hyperspectral technology and machine learning. J. Chin. Agric. Mech. 2019, 40, 71–77.
- Liu, Y.; Chen, X.; Wang, Z.F.; Ward, R.K.; Wang, X.S. Deep learning for pixel-level image fusion: Recent advances and future prospects. Inf. Fusion 2018, 42, 158–173.
- Sladojevic, S.; Arsenovic, M.; Anderla, A.; Culibrk, D. Deep neural networks based recognition of plant diseases by leaf image classification. Comput. Intell. Neurosci. 2016, 2016.
- Singh, A.; Ganapathysubramanian, B.; Singh, A.K.; Sarkar, S. Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci. 2016, 21, 110–124.
- Liu, H.; Coquin, D.; Valet, L.; Cerutti, G. Leaf species classification based on a botanical shape sub-classifier strategy. In Proceedings of the 2014 22nd International Conference on Pattern Recognition, Stockholm, Sweden, 24–28 August 2014; pp. 24–28.




