
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Gelan Ayana | + 3638 word(s) | 3638 | 2021-02-24 06:49:17 | | | |
| 2 | Vivi Li | + 1 word(s) | 3639 | 2021-02-25 03:11:12 | | |
Video Upload Options
Transfer learning is a machine learning approach that reuses a learning method developed for a task as the starting point for a model on a target task. The goal of transfer learning is to improve performance of target learners by transferring the knowledge contained in other (but related) source domains. As a result, the need for large numbers of target-domain data is lowered for constructing target learners. Due to this immense property, transfer learning techniques are frequently used in ultrasound breast cancer image analyses. In this study, we focus on transfer learning methods applied on ultrasound breast image classification and detection from the perspective of transfer learning approaches, pre-processing, pre-training models, and convolutional neural network (CNN) models. Finally, comparison of different works is carried out, and challenges—as well as outlooks—are discussed.
1. Introduction
Breast cancer is the second leading cause of death in women; 12.5% of women from different societies worldwide are diagnosed with breast cancer [1]. According to previous studies, early detection of breast cancer is crucial because it can contribute to up to a 40% decrease in mortality rate [2][3]. Currently, the ultrasound imaging technique has emerged as a popular imaging modality for the diagnoses of breast cancer, especially in young women with dense breasts [4]. This is because ultrasound (US) imaging is a non-invasive procedure and it can efficiently capture tissue properties [5][6][7]. Studies have shown that the false negative recognition rate in other breast diagnosis methods, such as biopsy and mammography (MG), decreased on using different modalities, such as US imaging [2]. Additionally, ultrasound imaging methods can be used to improve the tumor detection rate by up to 17% during breast cancer diagnoses [6]. Furthermore, the number of non-essential biopsies can be decreased by approximately 40%, thereby reducing medication costs [5]. An additional benefit of ultrasound imaging is that it uses non-ionizing radiation, which does not negatively affect health and requires relatively simple technology [7]. Therefore, ultrasound scanners are cheaper and more portable than mammography [5][6][7][8]. However, ultrasonic systems are not a standalone modality for breast cancer diagnoses [6][7]; instead, they are integrated with mammography and histological observations to validate results [8]. To improve the diagnostic capacity of ultrasound imaging, several studies have employed existing technologies [9]. Machine learning has solved many of the problems associated with ultrasound in terms of the classification, detection, and segmentation of breast cancer, such as false positive rates, limitation in indicating changes caused by cancer, lower applicability for treatment monitoring, and subjective judgments [10][11][12]. However, many machine learning methods perform well only under a common assumption, i.e., the training and test data are obtained from the same feature space and have the same distribution [13]. When the distribution changes, most numerical values of the models need to be constructed from scratch using newly collected training data [11][12][13]. In medical applications, including breast ultrasound imaging, it is difficult to collect the required training data and construct models in this manner [14]. Thus, it is advisable to minimize the need and effort required for acquiring the training data [13][14]. In such scenarios, transfer learning from one task to the target task would be desirable [15]. Transfer learning enables the use of a model previously trained on another domain as the target for learning [16]. Thus, it reduces the need and effort required to collect additional training data for learning [11][10][12][13][14][15][16].
Transfer learning is based on the principle that previously learned knowledge can be exceptionally implemented to solve new problems in a more efficient and effective manner [17][18]. Thus, transfer learning requires established machine learning approaches that retain and reuse previously learned knowledge [19][20][21]. Transfer learning was recently applied to breast cancer imaging in 2016, following the emergence of several convolutional neural network (CNN) models, including AlexNet, VGGNet, GoogLeNet, ResNet, and Inception, to solve visual classification tasks in natural images that are trained on natural image database such as ImageNet [22]. The first application of transfer learning to breast cancer imaging was reported in 2016 by Hyunh et al., where they assessed the performance achieved by using features transferred from pre-trained deep CNNs for classifying breast cancer through computer-aided diagnosis (CADx) [23]. Following this, Byra et al. published a paper where they proposed a neural transfer learning approach for breast lesion classification through ultrasound [24]. Shortly after this, Yap et al. [25] published their work, which proposed the use of deep neural learning methods for breast cancer detection; they studied three different methods—a patch-based LeNet approach, a U-Net model, and a transfer learning method—with a pre-trained fully convolutional network, AlexNet. Following these works, a large number of articles have been published in the area of applying transfer learning for breast ultrasound imaging [26][27][28][29].
2. Transfer Learning
2.1. Overview of Transfer Learning
Transfer learning is a popular approach for building machine learning models without concerns about the amount of available data [30]. Training a deep model may require a significant amount of data and computational resources; however, transfer learning can help address this issue. In many cases, a previously established model can be adapted to other problems [31] via transfer learning. For instance, it is possible to use a model that has been trained for one task, such as classifying cell types, and then fine-tuning it to accomplish another task, such as classifying tumors. Transfer learning is a particularly indispensable approach in tasks related to computer vision. Studies on transfer learning have shown [31][32][33] that features learned from significantly large image sets such as ImageNet are highly transferable to a variety of image recognition tasks. There are two approaches to transferring knowledge from one model to another. The popular approach is to change the last layer of the previously trained model and replace it with a randomly initialized one [34]. Following this, only the parameters in the top layer are trained for the new task, whereas all other parameters remain fixed. This method can be considered to be the application of the transferred model as a feature extractor [35], because the fixed portion acts as a feature extractor (Figure 1), while the top layer acts as a traditional, fully connected neural network layer without any special assumptions regarding the input [34][35]. This approach works better if the data and tasks are similar to the data and task on which the original model was trained. In cases where there is limited data to train a model for the target task, this type of transfer learning might be the only option to train a model without overfitting, because having fewer parameters to train also reduces the risk of overfitting [36]. In cases where more data is available for training, which is rare in medical settings, it is possible to unfreeze transferred parameters and train the entire network [34][35][36][37]. In this case, essentially, the initial values of the parameters are transferred [37]. The task of initializing the weights using a pre-trained model instead of initializing them randomly can provide the model with a favorable beginning and improve the rate of convergence [36][37] and fine-tuning. To preserve the initialization from pre-training, it is common practice to lower the learning rate by one order of magnitude [38][39]. To prevent changing the transferred parameters too early, it is customary to start with frozen parameters [40][41][42][43][44], train only randomly initialized layers until they converge, and then unfreeze all parameters and fine-tune (Figure 1) the entire network. Transfer learning is particularly useful when there is a limited amount of data for one task and a large volume of data for another similar task, or when there exists a model that has already been trained on such data [45]. However, even if there is sufficient data for training a model from scratch and the tasks are not related, initializing the parameters using a pre-trained model is still better than random initialization [46].
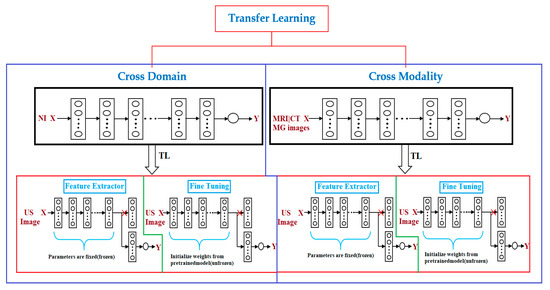
Figure 1. Transfer learning (TL) methods. There are two types of transfer learning used for breast cancer diagnosis via ultrasound imaging, depending on the source of pre-training data: cross-domain (model pre-trained on natural images is used) and cross-modal (model pre-trained on medical images is used). These two transfer learning approaches are feature extractors (convolution layers are used as a frozen feature extractor to match with a new task such as breast cancer classification) and fine-tuning (where instead of freezing convolution layers of the well-trained convolutional neural network (CNN) model, their weights are updated during the training process). X, input; Y, output; NI, natural image; MRI, magnetic resonance imaging; MG, mammography; CT, computed tomography; US, ultrasound.
2.2. Advantages of Transfer Learning
The main advantages of transfer learning include reducing training time, providing better performance for neural networks, and requiring limited data [47][48][49][50]. In neural networks trained on a large set of images, the early layer parameters resemble each other regardless of the specific task they have been trained on [16][47]. For example, CNNs tend to learn edges, textures, and patterns in the first layers [31], and these layers capture the features that are broadly useful for analyzing the natural images [47]. Features that detect edges, corners, shapes, textures, and different types of illuminants can be considered as generic feature extractors and can be used in many different types of settings [30][31][32][33]. The closer we get to the output, the more specific features the layers tend to learn [48][49][50]. For example, the last layer in a network that has been trained for classification would be highly specific to that classification task [49]. If the model was trained to classify tumors, one unit would respond only to the images of a specific tumor [23][24][25][26][28][27]. Transferring all layers except the top layer is the most common type of transfer learning [17][18][19][20]. Generally, it is possible to transfer the first n layers from a pre-trained model to a target network and randomly initialize the rest [51]. Technically, the transferred part does not have to be the first layer; if the tasks are similar, the type of input data is slightly different [21]. It is also possible to transfer the last layers [33]. For example, consider a tumor recognition model that has been trained on gray scale images and that the target is to build a tumor recognition model that inputs images that are colored in addition to gray scale data. Given that significant amounts of the data are not available to train a new model from scratch, it may be effective to transfer the latter layers and re-train the early ones [52][53]. Therefore, transfer learning is useful in the case where there is insufficient data for a new domain that is to be handled by a neural network and there exists a large pre-existing data pool that can be transferred to a target problem [47][48][49][50][51][52][53]. Transfer learning facilitates the building of a solid machine learning model with comparatively smaller training data because the model is already trained [53]. This is especially valuable in medical image processing because most of the time, data annotating persons are required to create large labeled datasets [24][25][26][28][27][29]. Furthermore, training time is minimized because it can reduce the time required to train a new deep neural network from the beginning in the case of complex target task [48][49].
2.3. Transfer Learning Approaches
Transfer learning has enabled researchers in the field of medical imaging, where there is a scarcity of data, to address the issue of small sample datasets and achieve better performance [13]. Transfer learning can be divided into two types—cross-domain and cross-modal transfer learning—based on whether the target and source data belong to the same domain [54][55]. Cross-domain transfer learning is a popular method for achieving a range of tasks in medical ultrasound image analyses [9]. In machine learning, the pre-training of models is conventionally accomplished on large sample datasets, and large training data ensure outstanding performance; however, this is far from reality, making the approach unsuitable in the medical imaging domain [56]. In the case of small training samples, the domain-specific models trained from scratch can work better [57][58][59][60] relative to transfer learning from a neural network model that has been pre-trained with large training samples in another domain, such as the natural image database of ImageNet. One of the reasons for this is that the gauging from the unprocessed image to the feature vectors used for a particular task, such as classification in the medical case, is sophisticated in the pre-trained case and requires a large training sample for improved generalization [58][59][60]. Instead, an exclusively designed small network will be ideal for limited training datasets that are usually experienced in medical imaging [13][58][59]. Furthermore, models trained on natural images are not suitable for medical images because medical images typically have low contrast and rich textures [61][62]. In such cases, cross-modal transfer learning performs better than cross-domain transfer learning [63]. In medical cases, especially in breast imaging, different modalities, such as magnetic resonance imaging (MRI), mammography (MG), computed tomography (CT), and ultrasound (US) are frequently used in the diagnostic workflow [63][64][65]. Mammography (i.e., X-ray) and ultrasound are the first-line screening methods for breast cancer examination, and it is trivial to collect large training samples compared to MRI and CT [66][67][68]. Breast MRI is a more costly, time-consuming method, and it is commonly used for screening high-risk populations, making it considerably difficult to acquire datasets and ground-truth annotation in the case of MRIs, as compared to ultrasound and mammograms [29]. In such instances, cross-modal transfer learning is an optimal approach [69][70]. A few experiments [29] have demonstrated the superiority of cross-modal transfer learning over cross-domain transfer learning for a given task in the case of smaller training datasets.
There are two popular approaches for transfer learning: feature extraction and fine-tuning [71] (Figure 1).
2.4. Pre-Training Model and Dataset
The most common pre-training models used for transfer learning in breast ultrasound are the VGG19, VGG16, AlexNet, and InceptionV3 models; VGG is the most common, followed by AlexNet and Inception, which are the least common. A comparison of the different pre-training models is not useful to determine the pre-training model that is better than the others for transfer learning in breast ultrasound [23][24][25][26][28][27][29]. However, one study [26], showed that Inception V3 outperforms VGG19, where the authors evaluated the impact of the ultrasound image reconstruction method on breast lesion classification using a neural transfer learning. In their study, a better overall classification performance was obtained for the classifier with the pre-training model using InceptionV3, which exhibited an AUC of 0.857. In the case of the VGG19 neural network, the AUC was 0.822.
Dataset usage for the pre-training of breast ultrasound transfer learning methods depends on whether cross-domain or cross-modal transfer learning methods are implemented [57][58][59][60]. In the case of cross-domain transfer learning, natural image datasets, such as ImageNet, are utilized as a pre-training dataset, whereas in the case of cross-modal transfer learning, datasets of MRI, CT, or MG images are utilized for pre-training the CNNs [23][24][25][26][28][27][29]. In the latter case, most researchers used their own data, although some used publicly available datasets. In breast ultrasound transfer learning, ImageNet is used, in most cases, as a pre-training dataset [23][24][25][26][28][27][29].
-
ImageNet: ImageNet is a large image database designed for use in image recognition [72][73][74]. It comprise more than 14 million images that have been hand-annotated to indicate the pictured objects. ImageNet is categorized into more than 20,000 categories with a typical category consisting of several images. The third-party image URLs repository of annotations is freely accessible directly from ImageNet, although ImageNet does not own the images.
2.5. Pre-Processing
The pre-processing required for applying transfer learning to breast ultrasound accomplishes two objectives [24][26]. The first is to compress the dynamic range of ultrasound signals to fit on the screen directly, and the second is to enlarge the dataset and reduce class imbalance. To achieve the first objective, [26] used a common method for ultrasound image analysis. First, the envelope of each raw ultrasound signal was calculated using the Hilbert transform. Next, the envelope was log-compressed, a specific threshold level was selected, and the log-compressed amplitude was mapped to the range of [0, 255]. In [24], Byra et.al used a matching layer where they proposed adjusting the grayscale ultrasound images to the pre-trained convolution neural network model instead of replicating grayscale images through the channels or changing the lower convolution layer of the CNN. Augmentation is used to achieve the second objective, which involves enlarging the dataset. Enlarging the amount of labeled data generally enhances the performance of CNN models [24][26]. Data augmentation is the process of synthetic data generation for training by producing variations in the original dataset [75][76]. For image data, the augmentation process involves different image manipulation techniques, such as rotation, translation, scaling, and flipping arrangements [76]. The challenging part for data augmentation are memory and computational constraints [77]. There are two popular data augmentation methods: online and offline data augmentation [78]. Online data augmentation is carried out on the fly during training, whereas offline data augmentation produces data in advance and stores it in memory [78]. The online approach saves storage but results in a longer training time, whereas the offline approach is faster in terms of training, although it consumes a large amount of memory [75][76][77][78].
2.6. Convolutional Neural Network
A CNN is a feed-forward neural network commonly used in ultrasound breast cancer image analysis [79]. The main advantage of the CNN is its accuracy in image recognition; however, it involves a high computational cost and requires numerous training data [80]. A CNN generally comprises an input layer, one or many convolution layers, pooling layers, and a fully connected layer [81]. The following are the most commonly used CNN models used for transfer learning with breast ultrasound images [79].
-
AlexNet: the AlexNet architecture is composed of eight layers. The first layers of AlexNet are the convolutional layers, and the next layer is a max-pooling layer for data dimension reduction [72][73][74]. AlexNet uses a rectified linear unit (ReLU) for the activation function, which offers faster training than other activation functions. The remaining three layers are the fully connected layers.
-
VGGNet: VGG16 was the first CNN introduced by the Visual Geometry Group (VGG); this was followed by VGG19; VGG16 and VGG19 becoming two excellent architectures on ImageNet [80]. VGGNet models afford better performance than AlexNet by superseding large kernel-sized filters with various small kernel-sized filters; thus, VGG16 and VGG19 comprise 13 and 16 convolution layers, respectively [79][80][82].
-
Inception: this is a GoogLeNet model focused on improving the efficiency of VGGNet from the perspective of memory usage and runtime without reducing performance accuracy [82][83][84][85]. To achieve this, it removes the activation functions of VGGNet that are iterative or zero [82]. Therefore, GoogLeNet came up with and added a module known as Inception, which approximates scattered connections between the activation functions [83]. Following InceptionV1, the architecture was improved in three subsequent versions [84][85]. InceptionV2 used batch normalization for training, and InceptionV3 introduced the factorization method to enhance the computational complexity of convolution layers. InceptionV4 brought about a similar comprehensive type of Inception-V3 architecture with a larger number of inception modules [85].
3. Discussion
It is evident that transfer learning has been incorporated in various application areas of ultrasound imaging analyses [15][16]. Although transfer learning methods have constantly been improving the existing capabilities of machine learning in terms of different aspects for breast ultrasound analyses, there still exists room for improvement [79][80][82][83][84][85].
In [26], the results depict several issues related to neural transfer learning. First, the image reconstruction procedures implemented in medical scanners should be considered. It is important to understand how medical images are acquired and reconstructed [75][76][77][78]. However, there is limited information regarding the image reconstruction algorithms implemented in ultrasound scanners. Typically, researchers involved in computer-aided diagnoses (CADx) system development agree that a particular system might not perform well on data acquired at another medical center using different scanners and protocols [83]. Their study [26] clearly shows that this issue might also be related to the CADx system being developed using data recorded in the same medical center.
In [24], the authors presented that the lack of demographic variations in race and ethnicity in the training data can negatively influence the detection and survival outcomes for underrepresented patient groups. They recommended that future works should seek to create a deep learning architecture with pre-training data collected from different imaging modalities. This pre-trained model can be useful for devising new automated detection systems based on medical imaging.
In [28], the performance of fine-tuning is demonstrated to be better than that of the feature extracting algorithm utilizing directly extracted CNN features; the authors obtained higher AUC values for the main dataset. However, the implementation of the fine-tuning approach is by far challenging and difficult, relative to the feature extracting approach [24][25][26][28][27][29]. It requires replacement of the fully connected layers in the initial CNN with custom layers [79]. Additionally, identifying the layers of the initial model that should be trained in the course of fine-tuning is difficult [79]. Moreover, to obtain enhanced performance on the test data, the parameters must be optimally selected, and constructing a fine-tuning algorithm is time consuming [80]. Furthermore, with a small dataset, fine-tuning may not be advisable, and it would be wiser to address such cases using a feature extraction approach [86][87].
Therefore, several important research issues need to be addressed in the area of transfer learning for breast cancer diagnoses via ultrasound imaging. In [29], the authors hypothesized that learning methods pre-trained on natural images, such as the ImageNet database, are not suitable for breast cancer ultrasound images because these are gray-level, low-contrast, and texture-rich images. They examined the implementation of a cross-modal fine-tuning approach, in which they used networks that were pre-trained on mammography (X-ray) images to classify breast lesions in MRI images. They found that cross-modal transfer learning with mammography and breast MRI would be beneficial to enhance the breast cancer classification performance in the face of limited training data. This work can be used to improve breast ultrasound imaging by applying cross-modal transfer learning from a network pre-trained on mammography or other modalities.
The phenomenon of color conversion is extensively employed in ultrasound image analyses [28]. In [28], the authors showed that color distribution is an important constraint that should be considered when attempting to efficiently utilize transfer learning with pre-trained models. With the application of color conversion, it was proved that one could make use of the pre-trained CNN more efficiently [79][80][82]. By utilizing the matching layer (ML), they were able to obtain better classification performance. The ML developed was proved to perform the same when using other datasets as well [28]. Thoroughly studying these applications and improving the performance of transfer learning should be another potential research direction.
References
- Mutar, M.T.; Goyani, M.S.; Had, A.M.; Mahmood, A.S. Pattern of Presentation of Patients with Breast Cancer in Iraq in 2018: A Cross-Sectional Study. J. Glob. Oncol. 2019, 5, 1–6.
- Coleman, C. Early Detection and Screening for Breast Cancer. Sem. Oncol. Nurs. 2017, 33, 141–155.
- Smith, N.B.; Webb, A. Ultrasound Imaging. In Introduction to Medical Imaging: Physics, Engineering and Clinical Applications, 6th ed.; Saltzman, W.M., Chien, S., Eds.; Cambridge University Press: Cambridge, UK, 2010; Volume 1, pp. 145–197.
- Gilbert, F.J.; Pinker-Domenig, K. Diagnosis and Staging of Breast Cancer: When and How to Use Mammography, Tomosynthesis, Ultrasound, Contrast-Enhanced Mammography, and Magnetic Resonance Imaging. Dis. Chest Breast Heart Vessels 2019, 2019–2022, 155–166.
- Jesneck, J.L.; Lo, J.Y.; Baker, J.A. Breast Mass Lesions: Computer-aided Diagnosis Models with Mammographic and Sonographic Descriptors. Radiology 2007, 244, 390–398.
- Feldman, M.K.; Katyal, S.; Blackwood, M.S. US artifacts. Radiographics 2009, 29, 1179–1189.
- Barr, R.; Hindi, A.; Peterson, C. Artifacts in diagnostic ultrasound. Rep. Med. Imaging 2013, 6, 29–49.
- Zhou, Y. Ultrasound Diagnosis of Breast Cancer. J. Med. Imag. Health Inform. 2013, 3, 157–170.
- Liu, S.; Wang, Y.; Yang, X.; Li, S.; Wang, T.; Lei, B.; Ni, D.; Liu, L. Deep Learning in Medical Ultrasound Analysis: A Review. Engineering 2019, 5, 261–275.
- Huang, Q.; Zhang, F.; Li, X. Machine Learning in Ultrasound Computer-Aided Diagnostic Systems: A Survey. BioMed Res. Int. 2018, 7, 1–10.
- Brattain, L.J.; Telfer, B.A.; Dhyani, M.; Grajo, J.R.; Samir, A.E. Machine learning for medical ultrasound: Status, methods, and future opportunities. Abdom. Radiol. 2018, 43, 786–799.
- Sloun, R.J.G.v.; Cohen, R.; Eldar, Y.C. Deep Learning in Ultrasound Imaging. Proc. IEEE 2020, 108, 11–29.
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359.
- Khoshdel, V.; Ashraf, A.; LoVetri, J. Enhancement of Multimodal Microwave-Ultrasound Breast Imaging Using a Deep-Learning Technique. Sensors 2019, 19, 4050.
- Day, O.; Khoshgoftaar, T.M. A survey on heterogeneous transfer learning. J. Big Dat. 2017, 4, 29.
- Weiss, K.; Khoshgoftaar, T.M.; Wang, D. A survey of transfer learning. J. Big Dat. 2016, 3, 1–9.
- Gentle Introduction to Transfer Learning. Available online: https://bit.ly/2KuPVMA (accessed on 10 November 2020).
- Taylor, M.E.; Kuhlmann, G.; Stone, P. Transfer Learning and Intelligence: An Argument and Approach. In Proceedings of the 2008 Conference on Artificial General Intelligence, Amsterdam, The Netherlands, 18–19 June 2008; pp. 326–337.
- Parisi, G.I.; Kemker, R.; Part, J.L.; Kanan, C.; Wermter, S. Continual lifelong learning with neural networks: A review. Neural Netw. J. Int. Neur. Net. Soci. 2019, 113, 54–71.
- Silver, D.; Yang, Q.; Li, L. Lifelong Machine Learning Systems: Beyond Learning Algorithms. In Proceedings of the AAAI Spring Symposium, Palo Alto, CA, USA, 25–27 March 2013; pp. 49–55.
- Chen, Z.; Liu, B. Lifelong Machine Learning. Syn. Lect. Art. Intel. Machn. Learn. 2016, 10, 1–145.
- Alom, M.Z.; Taha, T.; Yakopcic, C.; Westberg, S.; Hasan, M.; Esesn, B.; Awwal, A.; Asari, V. The History Began from AlexNet: A Comprehensive Survey on Deep Learning Approaches. arXiv 2018, arXiv:abs/1803.01164.
- Huynh, B.; Drukker, K.; Giger, M. MO-DE-207B-06: Computer-Aided Diagnosis of Breast Ultrasound Images Using Transfer Learning From Deep Convolutional Neural Networks. Int. J. Med. Phys. Res. Prac. 2016, 43, 3705–3705.
- Byra, M.; Galperin, M.; Ojeda-Fournier, H.; Olson, L.; O’Boyle, M.; Comstock, C.; Andre, M. Breast mass classification in sonography with transfer learning using a deep convolutional neural network and color conversion. Med. Phys. 2019, 46, 746–755.
- Yap, M.H.; Pons, G.; Marti, J.; Ganau, S.; Sentis, M.; Zwiggelaar, R.; Davison, A.K.; Marti, R.; Moi Hoon, Y.; Pons, G.; et al. Automated Breast Ultrasound Lesions Detection Using Convolutional Neural Networks. IEEE J. Biomed. Health Inform. 2018, 22, 1218–1226.
- Byra, M.; Sznajder, T.; Korzinek, D.; Piotrzkowska-Wroblewska, H.; Dobruch-Sobczak, K.; Nowicki, A.; Marasek, K. Impact of Ultrasound Image Reconstruction Method on Breast Lesion Classification with Deep Learning. arXiv 2018, arXiv:abs/1804.02119.
- Yap, M.H.; Goyal, M.; Osman, F.M.; Martí, R.; Denton, E.; Juette, A.; Zwiggelaar, R. Breast ultrasound lesions recognition: End-to-end deep learning approaches. J. Med. Imaging 2019, 6, 1–7.
- Hijab, A.; Rushdi, M.A.; Gomaa, M.M.; Eldeib, A. Breast Cancer Classification in Ultrasound Images using Transfer Learning. In Proceedings of the 2019 Fifth International Conference on Advances in Biomedical Engineering (ICABME), Tripoli, Lebanon, 17–19 October 2019; pp. 1–4.
- Hadad, O.; Bakalo, R.; Ben-Ari, R.; Hashoul, S.; Amit, G. Classification of breast lesions using cross-modal deep learning. IEEE 14th Intl. Symp. Biomed. Imaging 2017, 1, 109–112.
- Transfer Learning. Available online: http://www.isikdogan.com/blog/transfer-learning.html (accessed on 20 November 2020).
- Chu, B.; Madhavan, V.; Beijbom, O.; Hoffman, J.; Darrell, T. Best Practices for Fine-Tuning Visual Classifiers to New Domains. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2016; pp. 435–442.
- Transfer Learning. Available online: https://cs231n.github.io/transfer-learning (accessed on 19 November 2020).
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? Adv. Neur. Inf. Proc. Sys. (NIPS). 2014, 27, 1–14.
- Huh, M.-Y.; Agrawal, P.; Efros, A.A.J.A. What makes ImageNet good for transfer learning? arXiv 2016, arXiv:abs/1608.08614.
- Li, Z.; Hoiem, D. Learning without Forgetting. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 2935–2947.
- Building Trustworthy and Ethical AI Systems. Available online: https://www.kdnuggets.com/2019/06/5-ways-lack-data-machine-learning.html (accessed on 15 November 2020).
- Overfit and Underfit. Available online: https://www.tensorflow.org/tutorials/keras/overfit_and_underfit (accessed on 10 November 2020).
- Handling Overfitting in Deep Learning Models. Available online: https://towardsdatascience.com/handling-overfitting-in-deep-learning-models-c760ee047c6e (accessed on 12 November 2020).
- Transfer Learning: The Dos and Don’ts. Available online: https://medium.com/starschema-blog/transfer-learning-the-dos-and-donts-165729d66625 (accessed on 20 November 2020).
- Transfer Learning & Fine-Tuning. Available online: https://keras.io/guides/transfer_learning/ (accessed on 2 November 2020).
- How the pytorch freeze network in some layers, only the rest of the training? Available online: https://bit.ly/2KrE2qK (accessed on 2 November 2020).
- Transfer Learning. Available online: https://colab.research.google.com/github/kylemath/ml4aguides/blob/master/notebooks/transferlearning.ipynb (accessed on 5 November 2020).
- A Comprehensive Hands-on Guide to Transfer Learning with Real-World Applications in Deep Learning. Available online: https://towardsdatascience.com/a-comprehensive-hands-on-guide-to-transfer-learning-with-real-world-applications-in-deep-learning-212bf3b2f27a (accessed on 3 November 2020).
- Transfer Learning with Convolutional Neural Networks in PyTorch. Available online: https://towardsdatascience.com/transfer-learning-with-convolutional-neural-networks-in-pytorch-dd09190245ce (accessed on 25 October 2020).
- Best, N.; Ott, J.; Linstead, E.J. Exploring the efficacy of transfer learning in mining image-based software artifacts. J. Big Dat. 2020, 7, 1–10.
- He, K.; Girshick, R.; Dollar, P. Rethinking ImageNet Pre-Training. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), New York, NY, USA, 27 October–2 November 2019; pp. 4917–4926.
- Neyshabur, B.; Sedghi, H.; Zhang, C.J.A. What is being transferred in transfer learning? arXiv 2020, arXiv:abs/2008.11687.
- Liu, L.; Chen, J.; Fieguth, P.; Zhao, G.; Chellappa, R.; Pietikäinen, M. From BoW to CNN: Two Decades of Texture Representation for Texture Classification. Int. J. Comp. Vis. 2019, 127, 74–109.
- Çarkacioglu, A.; Yarman Vural, F. SASI: A Generic Texture Descriptor for Image Retrieval. Pattern Recogn. 2003, 36, 2615–2633.
- Yan, Y.; Ren, W.; Cao, X. Recolored Image Detection via a Deep Discriminative Model. IEEE Trans. Inf. Forensics Sec. 2018, 7, 1–7.
- Imai, S.; Kawai, S.; Nobuhara, H. Stepwise PathNet: A layer-by-layer knowledge-selection-based transfer learning algorithm. Sci. Rep. 2020, 10, 1–14.
- Zhao, Z.; Zheng, P.; Xu, S.; Wu, X. Object Detection with Deep Learning: A Review. IEEE Trans. Neur. Net. Learn. Sys. 2019, 30, 3212–3232.
- Transfer Learning (C3W2L07). Available online: https://www.youtube.com/watch?v=yofjFQddwHE&t=1s (accessed on 3 November 2020).
- Zhang, J.; Li, W.; Ogunbona, P.; Xu, D. Recent Advances in Transfer Learning for Cross-Dataset Visual Recognition: A Problem-Oriented Perspective. ACM Comput. Surv. 2019, 52, 1–38.
- Nguyen, D.; Sridharan, S.; Denman, S.; Dean, D.; Fookes, C.J.A. Meta Transfer Learning for Emotion Recognition. arXiv 2020, arXiv:abs/2006.13211.
- Schmidt, J.; Marques, M.R.G.; Botti, S.; Marques, M.A.L. Recent advances and applications of machine learning in solid-state materials science. NPJ Comput. Mater. 2019, 5, 1–36.
- D’souza, R.N.; Huang, P.-Y.; Yeh, F.-C. Structural Analysis and Optimization of Convolutional Neural Networks with a Small Sample Size. Sci. Rep. 2020, 10, 1–13.
- Rizwan I Haque, I.; Neubert, J. Deep learning approaches to biomedical image segmentation. Inform. Med. Unlocked 2020, 18, 1–12.
- Azizi, S.; Mousavi, P.; Yan, P.; Tahmasebi, A.; Kwak, J.T.; Xu, S.; Turkbey, B.; Choyke, P.; Pinto, P.; Wood, B.; et al. Transfer learning from RF to B-mode temporal enhanced ultrasound features for prostate cancer detection. Int. J. Comp. Assist. Radiol. Surg. 2017, 12, 1111–1121.
- Amit, G.; Ben-Ari, R.; Hadad, O.; Monovich, E.; Granot, N.; Hashoul, S. Classification of breast MRI lesions using small-size training sets: Comparison of deep learning approaches. Proc. SPIE 2017, 10134, 1–6.
- Tajbakhsh, N.; Jeyaseelan, L.; Li, Q.; Chiang, J.N.; Wu, Z.; Ding, X. Embracing imperfect datasets: A review of deep learning solutions for medical image segmentation. Med. Image Anal. 2020, 63, 1–30.
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629.
- Calisto, F.M.; Nunes, N.; Nascimento, J. BreastScreening: On the Use of Multi-Modality in Medical Imaging Diagnosis. arXiv 2020, arXiv:2004.03500v2.
- Evans, A.; Trimboli, R.M.; Athanasiou, A.; Balleyguier, C.; Baltzer, P.A.; Bick, U. Breast ultrasound: Recommendations for information to women and referring physicians by the European Society of Breast Imaging. Insights Imaging 2018, 9, 449–461.
- Mammography in Breast Cancer. Available online: https://bit.ly/2Jyf8pl (accessed on 20 November 2020).
- Eggertson, L. MRIs more accurate than mammograms but expensive. CMAJ 2004, 171, 840.
- Salem, D.S.; Kamal, R.M.; Mansour, S.M.; Salah, L.A.; Wessam, R. Breast imaging in the young: The role of magnetic resonance imaging in breast cancer screening, diagnosis and follow-up. J. Thorac. Dis. 2013, 5, 9–18.
- A Literature Review of Emerging Technologies in Breast Cancer Screening. Available online: https://bit.ly/37Ccmas (accessed on 20 October 2020).
- Li, W.; Gu, S.; Zhang, X.; Chen, T. Transfer learning for process fault diagnosis: Knowledge transfer from simulation to physical processes. Comp. Chem. Eng. 2020, 139, 1–10.
- Zhong, E.; Fan, W.; Yang, Q.; Verscheure, O.; Ren, J. Cross Validation Framework to Choose amongst Models and Datasets for Transfer Learning. In Proceedings of the Machine Learning and Knowledge Discovery in Databases, Berlin, Heidelberg, Germany, 12–15 July 2010; pp. 547–562.
- Baykal, E.; Dogan, H.; Ercin, M.E.; Ersoz, S.; Ekinci, M. Transfer learning with pre-trained deep convolutional neural networks for serous cell classification. Multimed. Tools Appl. 2020, 79, 15593–15611.
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252.
- Deng, J.; Dong, W.; Socher, R.; Li, L.; Kai, L.; Li, F.-F. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255.
- Khan, A.; Sohail, A.; Zahoora, U.; Qureshi, A.S. A survey of the recent architectures of deep convolutional neural networks. Artif. Intell. Rev. 2020, 53, 5455–5516.
- Mikołajczyk, A.; Grochowski, M. Data augmentation for improving deep learning in image classification problem. In Proceedings of the 2018 International Interdisciplinary PhD Workshop (IIPhDW), Swinoujście, Poland, 9–12 May 2018; pp. 117–122.
- Ma, B.; Wei, X.; Liu, C.; Ban, X.; Huang, H.; Wang, H.; Xue, W.; Wu, S.; Gao, M.; Shen, Q.; et al. Data augmentation in microscopic images for material data mining. NPJ Comput. Mat. 2020, 6, 1–14.
- Kamycki, K.; Kapuscinski, T.; Oszust, M. Data Augmentation with Suboptimal Warping for Time-Series Classification. Sensors 2019, 20, 95.
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Dat. 2019, 60, 1–48.
- Schmidhuber, J. Deep learning in neural networks: An overview. Neur. Net. 2015, 61, 85–117.
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556.
- Morid, M.A.; Borjali, A.; Del Fiol, G. A scoping review of transfer learning research on medical image analysis using ImageNet. Comput. Biol. Med. 2021, 128, 10–15.
- Szegedy, C.; Wei, L.; Yangqing, J.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9.
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the 32nd International Conference on Machine Learning, Lille, France, 20–23 February 2015; pp. 448–456.
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), City of Las Vegas, NY, USA, 27–30 June 2016; pp. 2818–2826.
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A.J.A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. arXiv 2017, arXiv:abs/1602.07261.
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J. Dig. Imaging 2019, 32, 582–596.
- Liu, L.; Ouyang, W.; Wang, X.; Fieguth, P.; Chen, J.; Liu, X.; Pietikäinen, M. Deep Learning for Generic Object Detection: A Survey. Int. J. Comput. Vis. 2020, 128, 261–318.




