
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Konstantina Vougogiannopoulou | + 1148 word(s) | 1148 | 2021-02-09 04:41:22 | | | |
| 2 | Catherine Yang | Meta information modification | 1148 | 2021-02-23 04:51:11 | | |
Video Upload Options
The ongoing pandemic of the severe acute respiratory syndrome (SARS), caused by the SARS-CoV-2 human coronavirus (HCoV), has brought the international scientific community before a state of emergency that needs to be addressed with intensive research for the discovery of pharmacological agents with antiviral activity.
1. Introduction
Coronaviruses (CoVs), are enveloped, positive-strand RNA viruses, with a genome of 27–33 kb, the largest in all RNA viruses. Their virion is spherical (approx. 125 nm diameter), with club-shaped spike-proteins (S protein) that stick out from the surface and result in a crown-like appearance of the enveloped virion[1]. There are seven known human CoVs (HCoVs), (Figure 1) two of which (HCoV-229E, HCoV-NL63) belong to the alpha genera of the subfamily Orthocoronavirinae[2] and the remaining five, HCoV-OC43, HCoV-HKU1, MERS-CoV, SARS-CoV, and SARS-CoV-2, belong to the beta genera. Most of the circulating HCoVs cause symptoms of common cold, although they occasionally can also cause severe or fatal disease. Three beta-CoVs, namely MERS-CoV, SARS-CoV, and SARS-CoV-2, emerged in the last 20 years causing several epidemics of acute respiratory illness associated with high mortality: 10% CFR for SARS CoV-1 and 34% for MERS-CoV[3][4]. The SARS-CoV-2-induced COVID-19 pandemic has caused more than one million deaths since the onset of the disease on 12 December. The genomic sequences of SARS-CoV and SARS-CoV-2 are 79.6% identical and their half-lives in aerosols and in plastic, metal, and cardboard surfaces are reportedly similar[5]. The comparatively far higher contagiousness and pandemic potential of SARS-CoV-2 are thought to reflect in part the substantial prevalence of undocumented contagious infections compared to the documented ones[5]. The contagiousness of the virus renders its containment difficult and the demand for prophylactic and therapeutic agents an utmost necessity that drives the scientific community in a massive screening effort. In this scenario, bioactive molecules from the vegetable kingdom are a source worthful to mine. The modern tools of natural products (NPs) chemistry (fast identification, dereplication, fast chemical profiling, in silico screening) and biological evaluation (high throughput in vitro screening assays, live infection assays, high throughput genomics and proteomics of host’s response to infection) provide ample means to explore plant biodiversity for discovery and/or development of NPs and small molecules (SMs) that can help cope with COVID-19 and here we summarize the efforts accomplished up to date.
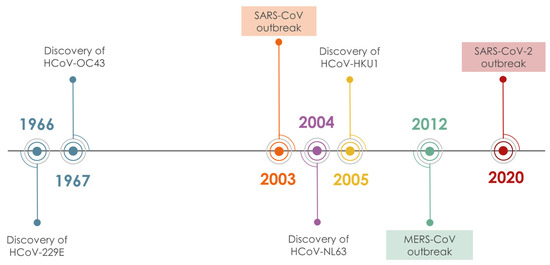
Figure 1. Timeline of HCoV discovery.
2. NPs with Anti-HCoV Potential
Before the emergence of SARS-CoV and MERS-CoV, the investigation of NPs as anti-HCoV agents was limited to only a few studies. The two epidemics have spurred however the interest in the discovery of anti-HCoV agents, including the investigation of agents active against the “common cold” HCoVs, 229E, NL63, OC43, and HKU1. Screening chemical libraries against target proteins and/or viral replication in cell-based assays have been extensively employed, resulting in the discovery of several antiviral natural products/small molecules (NPs/SMs)[6][7][8][9]. It was found that NPs/SMs are a rich source of drug potential leads against coronaviruses due to their pronounced structural diversity and complexity. Several compilations of pre-2020 findings on NPs/SMs with activity against coronaviruses were recently reported, some of which could serve as leads for the development of new drugs[10][11][12][13][14][15][16][17][18][19][20][21]. Among the identified molecules the majority is represented by NPs (Figure 2a) and most of them have been found to act as inhibitors of SARS-CoV (Figure 2b). Among them, the most numerous classes are flavonoids, triterpenes, and alkaloids (Figure 2c), which is not surprising since in these classes reside the greatest number of NPs described as endowed with inhibitory activity against replication of multiple different viruses. In addition, flavonoids are a class of NPs present in almost every plant species and are strongly represented in various NP-based chemical libraries available for screening.
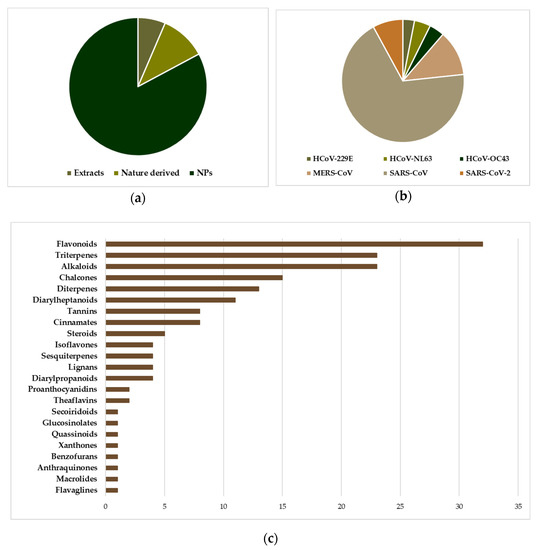
Figure 2. The position of natural products implicated in anti-HCoV research. (a) Distribution of entities (extracts, NPs, Nature derived molecules) subjected to anti-HCoV assays (cell-based or viral enzyme inhibition), in a total of 326 experiments corresponding to 197 entities. “Nature derived” refers to synthetic/semisynthetic NPs with a basic core inspired by NPs. (b) The prevalence of SARS-CoV testing among several NP entities. (c) The distribution of the 168 pure NPs tested in various anti-HCoV assays, with respect to the chemical category.
In the majority of studies, the initial screening involves the addition of the compound or extract to be tested in the normal cell culture, and a subsequent inoculation with the desired viral strain. The cytopathic morphology of the cells is evaluated under a microscope, while the cytotoxicity of the compound/extract is evaluated in normal cells and compared. Compounds with low cytotoxicity are then typically evaluated in multiple concentrations for their ability to inhibit viral replication (EC50) and host cell growth (CC50), and the selectivity index (SI, CC50/EC50 ratio) is used to identify the lead compounds, where higher SI refers to compounds more active than toxic. To a first step in investigating the molecular mechanisms underlying the antiviral activity, interactions of the lead compounds with identified targets, such as viral proteases, host proteases, or viral spike (S) proteins, are evaluated. Due to the additive complexity of the host/virus system, it is rather difficult to correlate cytopathic effects (CPE) to specific target interaction/inhibition, and this is the case for most natural products tested for their anti-HCoV potential, where extensive mechanistic studies are limited.
3. Conclusions
Natural products have played an active and important role in drug discovery up until today. Nevertheless, their valorization as antiviral agents remains limited. It is indicative that antiviral NP research has peaked only around imminent threats, as happened with SARS-CoV. Our review of the bibliography covering the study of NPs against HCoVs, revealed that there are many possibilities in examining more thoroughly available NPs in order to discover new antiviral agents. NP databases and NP libraries with physical samples available for bioactivity screening may have shortened the time needed for a compound to reach bioactivity evaluation, but as shown from the literature survey, tested compounds tend to revolve around very common and not very diverse structures. In terms of anti HCoV activity and NPs, the available literature is limited in order to draw sound conclusions about structure-activity relationships. Nevertheless, the data show that there is a trend for alkaloids, triterpenoids/triterpene saponins, and polyhydroxylated flavonoids. HCoVs and NPs research has been fragmented up to this point, driven by states of emergency, such as the emergence of SARS-CoV in 2002, and the current threat of SARS-CoV-2. Although there are a number of published results, there is a lack of systematic investigation of NPs showing promising activity, and this needs to be addressed by research efforts defining the mechanism of action of these compounds. Under this scope, compounds based on NPs scaffolds with higher potency, more favorable physicochemical properties, and diminished toxicity, can be designed, thus providing a multidisciplinary approach that antiviral discovery needs. NPs offer great chemodiversity that needs to be further exploited, especially under the current pressure of this global pandemic.
References
- Maier, H.J.; Bickerton, E.; Britton, P. Coronaviruses: Methods and Protocols; Humana Press: Heidelberg, Germany, 2015; pp. 1–282. ISBN 9781493924387.
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544.
- Memish, Z.A.; Perlman, S.; Van Kerkhove, M.D.; Zumla, A. Middle East respiratory syndrome. Lancet 2020, 395, 1063–1077.
- Cheng, V.C.C.; Lau, S.K.P.; Woo, P.C.Y.; Yuen, K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007, 20, 660–694. [
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567.
- Xiong, B.; Gui, C.-S.; Xu, X.-Y.; Luo, C.; Chen, J.; Luo, H.-B.; Chen, L.-L.; Li, G.-W.; Sun, T.; Yu, C.-Y.; et al. A 3D model of SARS_CoV 3CL proteinase and its inhibitors design by virtual screening. Acta Pharm. Sin. 2003, 24, 497–504+619.
- Pillaiyar, T.; Manickam, M.; Namasivayam, V.; Hayashi, Y.; Jung, S.H. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: Peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016, 59, 6595–6628.
- Wu, C.-Y.; Jan, J.-T.; Ma, S.-H.; Kuo, C.-J.; Juan, H.-F.; Cheng, Y.-S.E.; Hsu, H.-H.; Huang, H.-C.; Wu, D.; Brik, A.; et al. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. USA 2004, 101, 10012–10017.
- Kao, R.Y.; Tsui, W.H.W.; Lee, T.S.W.; Tanner, J.A.; Watt, R.M.; Huang, J.-D.; Hu, L.; Chen, G.; Chen, Z.; Zhang, L.; et al. Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem. Biol. 2004, 11, 1293–1299.
- Islam, M.T.; Sarkar, C.; El-Kersh, D.M.; Jamaddar, S.; Uddin, S.J.; Shilpi, J.A.; Mubarak, M.S. Natural products and their derivatives against coronavirus: A review of the non-clinical and pre-clinical data. Phyther. Res. 2020, 34, 2471–2492.
- Yang, Y.; Islam, M.S.; Wang, J.; Li, Y.; Chen, X. Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): A review and perspective. Int. J. Biol. Sci. 2020, 16, 1708–1717.
- Al-Hatamleh, M.A.I.; Hatmal, M.M.; Sattar, K.; Ahmad, S.; Mustafa, M.Z.; Bittencourt, M.D.C.; Mohamud, R. Antiviral and Immunomodulatory Effects of Phytochemicals from Honey against COVID-19: Potential mechanisms of action and future Directions. Molecules 2020, 25, 5017.
- Boozari, M.; Hosseinzadeh, H. Natural products for COVID-19 prevention and treatment regarding to previous coronavirus infections and novel studies. Phyther. Res. 2020.
- Liu, Y.; Liang, C.; Xin, L.; Ren, X.; Tian, L.; Ju, X.; Li, H.; Wang, Y.; Zhao, Q.; Liu, H.; et al. The development of Coronavirus 3C-Like protease (3CLpro) inhibitors from 2010 to 2020. Eur. J. Med. Chem. 2020, 206, 112711.
- Tiwari, V.; Beer, J.C.; Sankaranarayanan, N.V.; Swanson-Mungerson, M.; Desai, U.R. Discovering small-molecule therapeutics against SARS-CoV-2. Drug Discov. Today 2020, 25, 1535–1544.
- Zhou, J.; Huang, J. Current Findings Regarding Natural Components with Potential Anti-2019-nCoV Activity. Front. Cell Dev. Biol. 2020, 8, 589.
- da Silva Antonio, A.; Wiedemann, L.S.M.; Veiga-Junior, V.F. Natural products’ role against COVID-19. RSC Adv. 2020, 10, 23379–23393.
- Khalifa, S.A.M.; Yosri, N.; El-Mallah, M.F.; Ghonaim, R.; Guo, Z.; Musharraf, S.G.; Du, M.; Khatib, A.; Xiao, J.; Saeed, A.; et al. Screening for natural and derived bio-active compounds in preclinical and clinical studies: One of the frontlines of fighting the coronaviruses pandemic. Phytomedicine 2020, 153311.
- Verma, S.; Twilley, D.; Esmear, T.; Oosthuizen, C.B.; Reid, A.M.; Nel, M.; Lall, N. Anti-SARS-CoV Natural Products with the Potential to Inhibit SARS-CoV-2 (COVID-19). Front. Pharmacol. 2020, 11, 561334.
- Adhikari, B.; Marasini, B.P.; Rayamajhee, B.; Bhattarai, B.R.; Lamichhane, G.; Khadayat, K.; Adhikari, A.; Khanal, S.; Parajuli, N. Potential roles of medicinal plants for the treatment of viral diseases focusing on COVID-19: A review. Phyther. Res. 2020.
- Wang, Z.; Yang, L. Turning the Tide: Natural Products and Natural-Product-Inspired Chemicals as Potential Counters to SARS-CoV-2 Infection. Front. Pharmacol. 2020, 11, 1013.




