
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jiansheng Huang | + 779 word(s) | 779 | 2020-04-22 09:47:07 | | | |
| 2 | Nicole Yin | -1 word(s) | 778 | 2020-05-11 09:55:46 | | | | |
| 3 | Nicole Yin | -5 word(s) | 773 | 2020-10-28 07:00:19 | | |
Video Upload Options
Single-strand RNA (ssRNA) viruses such as the coronavirus family replicate the virus genomes by taking advantage of host cells. For example, after coronavirus approaches the ribosome of the epithelial cells or other host cells, it uses the ribosome of the host cell to replicate polyproteins. The replication and subsequent processes of precursor polyproteins can occur in the epithelial cells. After the coronavirus’ polyproteins are expressed, two enzymes — specifically, coronavirus main proteinase (3CLpro) and the papain-like protease (PLpro) — are thought to be involved in cleaving the polyproteins into smaller products used for replicating new viruses. In order to generate the daughter RNA genome, the coronavirus expresses an RNA-dependent RNA polymerase (RdRp), which is a crucial replicase that catalyzes the synthesis of a complementary RNA strand using the virus RNA template
1. Molecular Mechanisms of Coronavirus Invasion
The coronavirus (CoV) family has a large homogeneous “spike protein”. This spike protein (S protein) is responsible for interacting with the host cells, such as the pulmonary and parabronchial epithelial cell, and helps the coronavirus get through the epithelial cell membrane[1]. In addition, the alveolar epithelial cells have abundant expression of angiotensin-converting enzyme 2 (ACE2), which is targeted by the virus. The recognition of ACE2 by the S protein of the virus enables the invasion of the coronavirus into the human circulation system[2]. Recent study demonstrates that ACE2 is the SARS-CoV-2 receptor, which is required for cell entry[3]. Single-strand RNA (ssRNA) viruses such as the coronavirus family replicate the virus genomes by taking advantage of host cells. For example, after coronavirus approaches the ribosome of the epithelial cells or other host cells, it uses the ribosome of the host cell to replicate polyproteins. The replication and subsequent processes of precursor polyproteins can occur in the epithelial cells[4]. After the coronavirus’ polyproteins are expressed, two enzymes — specifically, coronavirus main proteinase (3CLpro) and the papain-like protease (PLpro) — are thought to be involved in cleaving the polyproteins into smaller products used for replicating new viruses[5]. In order to generate the daughter RNA genome, the coronavirus expresses an RNA-dependent RNA polymerase (RdRp), which is a crucial replicase that catalyzes the synthesis of a complementary RNA strand using the virus RNA template as shown in Figure 1[6].
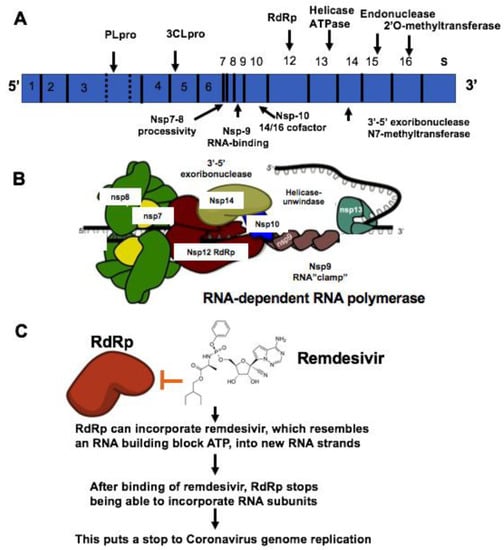
Figure 1. Mechanisms of remdesivir to inhibit RNA-dependent RNA polymerase (RdRp). (A) The genome composition model of single strand RNA (ssRNA) of coronavirus. (B) The RNA-dependent RNA polymerase RdRp mediated RNA replication during coronavirus infection. (C) Remdesivir functions as the ATP analog to inhibit RdRp.
2. Factors Involved in Transcription and Release of Coronavirus Particles
Although genome replication and transcription are well known to be regulated by the viral RdRp, several host factors have been implicated in this process. RNA chaperones are usually nonspecific nucleic acid binding proteins, which have long disordered structures that promote RNA molecules to adjust conformational changes. For example, coronavirus nucleoproteins (N protein) have RNA chaperone activity and function as an RNA chaperone, which could help template switching[7][8][9]. In addition, recent studies demonstrate that glycogen synthase kinase 3 (GSK3) phosphorylates the N protein of SARS-CoV and further inhibition of GSK3 can effectively inhibit viral replication in Vero E6 cells infected with SARS-CoV[10]. Furthermore, heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) is involved in the pre-mRNA splicing in the nucleus and translation regulation in the host cells. Importantly, it has been shown that the nucleocapsid protein of SARS-CoV had binding ability to human hnRNP A1 with high affinity by using kinetic analyses with a surface plasmon resonance (SPR) approach. These studies suggest that hnRNPA1 is able to bind to SARS-CoV N protein to form a replication/transcription complex and control viral RNA synthesis[11].
In addition, several virus proteins and host factors are essential for the assembly and release of coronavirus. Homotypic interaction of M protein serves as the scaffold for the virus assembly and morphogenesis in the infected cells, specifically, both the interaction between the membrane (M) and S protein and the interaction between M and N protein promote the recruitment of structural components to the assembly location of host cells[12][13]. For example, both envelope (E) protein and N proteins are required to be co-expressed with M protein for the formation and release of virus-like particles (VLPs) after transfection of Vero E6 cells. Two crucial structural proteins, the M protein and E protein, play important roles in the coronavirus assembly. In addition, the E protein is involved in particle assembly by binding with M and further inducing membrane curvature[14]. Subsequently, coronavirus particles can be budded into the ER-Golgi intermediate compartment (ERGIC) of host cells, then trafficked in a smooth-wall vesicle and transported through the secretory pathway for assembly and release by exocytosis[15][16].
References
- Shuai Xia; Yun Zhu; Meiqin Liu; Qiaoshuai Lan; Wei Xu; Yanling Wu; TianLei Ying; Shuwen Liu; Zhengli Shi; Shibo Jiang; et al.Lu Lu Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cellular & Molecular Immunology 2020, null, 1-3, 10.1038/s41423-020-0374-2.
- Sandrine Belouzard; Victor C. Chu; Gary R. Whittaker; Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites.. Proceedings of the National Academy of Sciences 2009, 106, 5871-6, 10.1073/pnas.0809524106.
- Markus Hoffmann; Hannah Kleine-Weber; Simon Schroeder; Nadine Krüger; Tanja Herrler; Sandra Erichsen; Tobias S. Schiergens; Georg Herrler; Nai-Huei Wu; Andreas Nitsche; et al.Marcel A. MüllerChristian DrostenStefan Pöhlmann SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271-280.e8, 10.1016/j.cell.2020.02.052.
- Markus Hoffmann; Hannah Kleine-Weber; Nadine Krüger; Marcel Müller; Christian Drosten; Stefan Pöhlmann; Marcel A Mueller; The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv 2020, null, null, 10.1101/2020.01.31.929042.
- Daniel Wrapp; Nianshuang Wang; Kizzmekia S. Corbett; Jory A. Goldsmith; Ching-Lin Hsieh; Olubukola Abiona; Barney S. Graham; Jason S. McLellan; Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260-1263, 10.1126/science.abb2507.
- Asher Mullard; Ebola outbreak prompts experimental drug rollout. Nature Reviews Drug Discovery 2018, 17, 460-460, 10.1038/nrd.2018.114.
- Sonia Zuñiga; Jazmina Libertad Gonzalez Cruz; Isabel Sola; Pedro A. Mateos-Gómez; Lorena Palacio; Luis Enjuanes; Coronavirus Nucleocapsid Protein Facilitates Template Switching and Is Required for Efficient Transcription. Journal of Virology 2009, 84, 2169-2175, 10.1128/jvi.02011-09.
- Kelly-Anne Spencer; Michael Dee; Paul Britton; Julian A. Hiscox; Role of phosphorylation clusters in the biology of the coronavirus infectious bronchitis virus nucleocapsid protein. Virology 2008, 370, 373-381, 10.1016/j.virol.2007.08.016.
- Sonia Zuñiga; Isabel Sola; Jose L. Moreno; Patricia Sabella; Juan Plana-Durán; Luis Enjuanes; Coronavirus nucleocapsid protein is an RNA chaperone. Virology 2007, 357, 215-227, 10.1016/j.virol.2006.07.046.
- C.-H. Wu; S.-H. Yeh; Y.-G. Tsay; Y.-H. Shieh; Chuan-Liang Kao; Y.-S. Chen; S.-H. Wang; T.-J. Kuo; Pei-Jer Chen; Pei-Jer Chen; et al. Glycogen Synthase Kinase-3 Regulates the Phosphorylation of Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Protein and Viral Replication. Journal of Biological Chemistry 2008, 284, 5229-5239, 10.1074/jbc.m805747200.
- Haibin Luo; Qing Chen; Jing Chen; Kaixian Chen; Xu Shen; Hualiang Jiang; The nucleocapsid protein of SARS coronavirus has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1. FEBS Letters 2005, 579, 2623-2628, 10.1016/j.febslet.2005.03.080.
- Mina Nakauchi; Hiroaki Kariwa; Yasuhiro Kon; Kentaro Yoshii; Akihiko Maeda; Ikuo Takashima; Analysis of severe acute respiratory syndrome coronavirus structural proteins in virus‐like particle assembly. Microbiology and Immunology 2008, 52, 625-630, 10.1111/j.1348-0421.2008.00079.x.
- Y. L. Siu; K. T. Teoh; J. Lo; C. M. Chan; F. Kien; N. Escriou; S. W. Tsao; John M. Nicholls; R. Altmeyer; J. S. M. Peiris; Roberto Bruzzone; Béatrice Nal; The M, E, and N Structural Proteins of the Severe Acute Respiratory Syndrome Coronavirus Are Required for Efficient Assembly, Trafficking, and Release of Virus-Like Particles. Journal of Virology 2008, 82, 11318-11330, 10.1128/jvi.01052-08.
- K. P. Lim; D. X. Liu; The Missing Link in Coronavirus Assembly. Journal of Biological Chemistry 2001, 276, 17515-17523, 10.1074/jbc.m009731200.
- To Sing Fung; Ding Xiang Liu; Human Coronavirus: Host-Pathogen Interaction.. Annual Review of Microbiology 2019, 73, 529-557, 10.1146/annurev-micro-020518-115759.
- Federica Brandizzi; Charles Barlowe; Organization of the ER-Golgi interface for membrane traffic control.. Nature Reviews Molecular Cell Biology 2013, 14, 382-92, 10.1038/nrm3588.




