Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | jean-marc corsi | -- | 4456 | 2024-03-15 13:32:10 | | | |
| 2 | Peter Tang | + 2 word(s) | 4458 | 2024-03-18 02:22:33 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Corne, A.; Adolphe, F.; Estaquier, J.; Gaumer, S.; Corsi, J. ATF4 Role during HIV-1 Replication. Encyclopedia. Available online: https://encyclopedia.pub/entry/56343 (accessed on 08 February 2026).
Corne A, Adolphe F, Estaquier J, Gaumer S, Corsi J. ATF4 Role during HIV-1 Replication. Encyclopedia. Available at: https://encyclopedia.pub/entry/56343. Accessed February 08, 2026.
Corne, Adrien, Florine Adolphe, Jérôme Estaquier, Sébastien Gaumer, Jean-Marc Corsi. "ATF4 Role during HIV-1 Replication" Encyclopedia, https://encyclopedia.pub/entry/56343 (accessed February 08, 2026).
Corne, A., Adolphe, F., Estaquier, J., Gaumer, S., & Corsi, J. (2024, March 15). ATF4 Role during HIV-1 Replication. In Encyclopedia. https://encyclopedia.pub/entry/56343
Corne, Adrien, et al. "ATF4 Role during HIV-1 Replication." Encyclopedia. Web. 15 March, 2024.
Copy Citation
Activating transcription factor 4 (ATF4) is a transcription factor known to regulate genes associated with the sensing of cellular stress such as amino acid deprival, protein misfolding, growth arrest, and cell death. Despite its key role at the crossroads of immune and stress responses, the precise impact of ATF4 during viral infections remains unclear. Thus, ATF4 has a dual role in promoting cell survival or cell death, but also in limiting infection or participating in viral replication.
ISR
AIDS
immunity
mitochondria
ER stress
UPR
1. Introduction
One of the main regulators of cellular homeostasis is activating transcription factor 4 (ATF4). This bZIP domain transcription factor plays a crucial role in both the integrated stress response (ISR) and the mitochondrial stress response (MSR) [1][2][3]. In reaction to oxidative stress, misfolded protein accumulation, or nutrient deprivation, ATF4 triggers the expression of genes involved in cellular processes participating in the control of autophagy or cell death. Furthermore, the role of ATF4 in cellular responses to stress is intimately linked to its dimerization partners such as the transcription factor ATF5, an ATF4 paralogue found in mammals, which belongs to the ATF4 bZIP-domain transcription factor family [4][5]. ATF5 is also strongly involved in stress responses and can be a target of ATF4. Thus, ATF4 and ATF5 are key cellular factors that integrate various stress signals.
2. HIV-1 Infection Regulates ATF4
2.1. ATF4 Is Up-Regulated during HIV-1 and SIV Infections
HIV-1 infection leads to the development of acquired immunodeficiency syndrome (AIDS) associated with the depletion of CD4+ T cells, mainly by apoptosis [6][7][8] which predicts further pathogenicity [9][10]. Of interest, the induction of ATF4 following HIV-1 infection has been observed in several in vitro models. It was first described in Jurkat T cells, in which ATF4 was up-regulated at both the transcript and protein levels 8 h post-infection and remained elevated 48 h later [11]. This increase in ATF4 transcript levels was further observed in primary human CD4+ T cells at day 5 post-infection [12]. In a model of HIV-1 latency, ATF4 transcript and protein levels are weakly detectable [13] but increased with viral reactivation [11][12][13]. In vivo, in monkeys infected with simian immunodeficiency virus (SIV), ATF4 transcripts are also up-regulated in the gut mucosa. This increase occurred during the acute phase of SIV infection, i.e., 1 to 2 weeks post-infection, but are not observed during the chronic phase [12]. Viral proteins released in the microenvironment of infected cells such as Tat has been proposed to be sufficient for inducing ATF4 gene expression through ER stress in non-infected cells [14]. Altogether, these observations show that ATF4 expression is differentially regulated during both the early and chronic phases of viral replication, in infected but also non-infected cells, which are close to the former, suggesting that ATF4 induction is related to viral stress and plays a potential role in the establishment of latency.
2.2. How Can HIV-1 Regulate ATF4?
2.2.1. HIV-1-Induced ISR/ATF4 Signaling
The ISR leads to a global translation blockade through the phosphorylation of eIF2α, which increases ATF4 translation [3]. Several kinases are responsible for eIF2α phosphorylation: double-stranded RNA-dependent protein kinase (PKR), PKR-like ER kinase (PERK), general control non-derepressible 2 (GCN2), heme-regulated eIF2α kinase (HRI), microtubule affinity-regulating kinase 2 (MARK2), and family with sequence similarity 69 member C (FAM69C) [3][15][16] (Figure 1). The role of eIF2α and its kinases during viral infection has been recently reviewed by Liu et al. [17]. The researchers focus on HIV-1 studies providing evidence that eIF2α phosphorylation by ISR kinases can be associated with ATF4 induction, and that a direct control of these kinases by HIV-1 proteins may affect ATF4 activity.
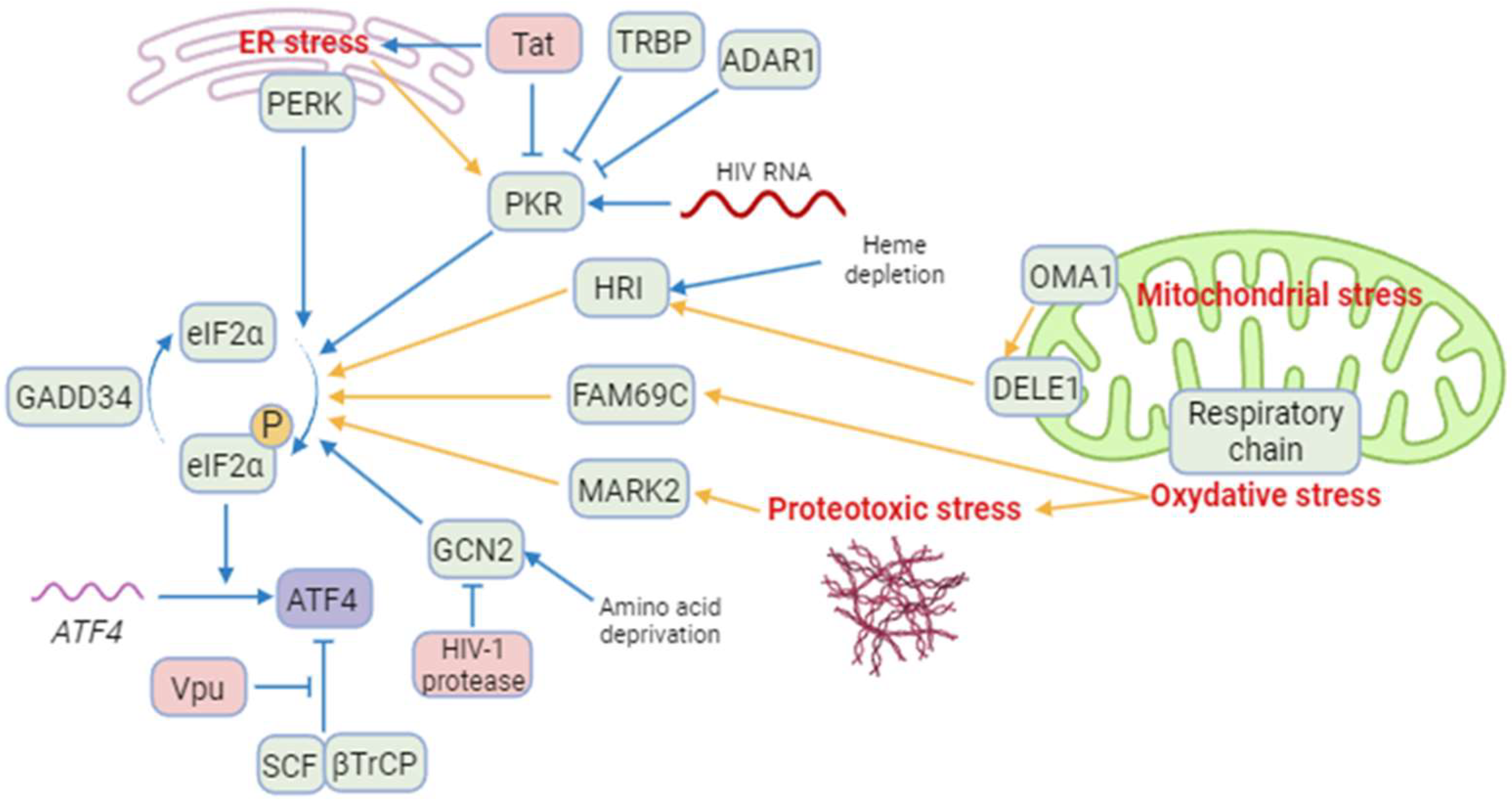
Figure 1. A model of regulation of ATF4 and integrated stress kinases during HIV-1 infection. Blue arrows correspond to interactions demonstrated by the literature in the context of HIV-1 infection. Yellow arrows correspond to reports in a different context. Besides ATF4, cellular proteins are indicated in green and HIV-1 proteins in red. ISR kinases can be activated by ER stress (PERK and PKR), HIV-1 RNA (PKR), heme depletion and cleavage of DELE1 by OMA1 after mitochondrial stress (HRI), amino acid deprival (GCN2), oxidative stress (FAM69C), and proteotoxic stress (MARK2). Several cellular and viral proteins modulate ISR kinases during HIV-1 infection, as TRBP and ADAR1 and the HIV-1 Tat protein preventing the phosphorylation of eIF2α and increasing ATF4 synthesis. The viral Vpu protein stabilizes ATF4 by opposing its ubiquitination by the SCF-βTrCP complex. Created with BioRender.com (accessed on 18 January 2024).
Among the ISR kinases, PKR is the best characterized viral nucleic acid sensor [18][19]. It is activated mainly by double-stranded RNAs (dsRNA) [20][21][22][23]. PKR regulates HIV-1 infection as it can bind the hairpin structure within the transactivation-response region of the HIV-1 genome [24]. While PKR contributes to ATF4 translation in response to endoplasmic reticulum (ER) stress [25], paradoxically, to the researchers' knowledge, no study reports the induction of ATF4 in the context of HIV-1 infection through an endogenous PKR/eIF2α pathway. A reason that may explain this lack of evidence is the rapid inhibition of PKR by multiple mechanisms during HIV-1 replication [24] (Figure 1): PKR activation is inhibited by high levels of dsRNA and by the direct binding of cellular proteins, including TAR RNA-binding protein (TRBP) and adenosine deaminase acting on RNA (ADAR1) [26][27][28][29][30][31][32]. Moreover, the viral Tat protein may counteract PKR activation by preventing PKR autophosphorylation. Tat also serves as a competitive inhibitor limiting PKR binding to eIF2α thanks to a sequence homology between one of its motifs and eIF2α [33][34][35].
Cells experiencing ER stress due to nutrient deprivation or viral replication may lead to the activation of PERK. Like many other viruses, HIV-1 is known to induce protein misfolding and ER stress [36]. Campestrini et al. have previously reported on the induction of ER unfolded protein response (UPR) genes such as PERK and ATF4, following the treatment of Jurkat cells with the HIV-1 Tat protein [14]. Additionally, PERK activation has been observed in Tat-treated human brain microvascular endothelial cells (HBMECs) and in HIV-1-infected CD4+ T cell lines [37][38]. While PERK is one of the sensors of the UPR with IRE-1 and ATF6, the functional role of PERK in ATF4 induction in response to HIV-1 infection is not fully demonstrated.
GCN2 is activated in response to amino acid deprival and metabolic dysfunction (Figure 1). These alterations are reported in plasma of HIV-1-infected patients and in the gut of SIV-infected monkeys [12][39][40][41][42]. Moreover, GCN2 is activated in vitro by HIV-1 or SIV infection [12][43] suggesting that metabolic disorders observed in HIV-1-infected patients could also lead to GCN2 activation. GCN2 phosphorylates the HIV-1 integrase, which reduces viral integration [44]. This property is not limited to HIV-1 since various integrases from other retroviruses are also recognized as substrates by GCN2 [44]. The anti-viral activity of GCN2 can be overcome by the HIV-1 protease that can cleave GCN2 [45]. Jiang et al. have reported that HIV-1-induced ATF4 transcription would result from the activation of the GCN2/ATF4 pathway [12]. Serum deprivation triggers HIV-1 replication of CD4+ T cells, correlating with the GCN2-mediated activation of ATF4, which is recruited to the HIV-1 long terminal repeat (LTR) to facilitate viral transcription.
Both heme/iron depletion and arsenite-induced oxidative stress activate HRI [3]. Arsenite-induced HRI activation increases viral protein production and the infection rate of reovirus [46]. It can also regulate the level of the viral factor Zta that plays a role in the lytic cycle of EBV [47][48]. HRI has been shown to be able to activate ATF4, triggering the expression of genes involved in autophagy [49][50][51] and oxidative-stress response including the antioxidant heme oxygenase-1 (HO-1) gene [51]. Different groups have reported a role of HO-1 in regulating viral replication [52][53] including HIV-1′s [54]. HO-1 dysfunction has been associated with neuronal diseases in people living with HIV-1 [55]. For instance, despite a link between HO-1 and viral replication, to date, no study has addressed the role of an HRI pathway regulating HIV-1 infection and replication.
Recently, two other kinases able to phosphorylate eIF2α have been characterized, namely MARK2 and FAM69C [15][16]. Both kinases are induced by proteotoxic stress. FAM69C KO mice-derived microglia displayed an indirect increase in inflammation, and HIV-1 binds MARK2 to control motor adaptor function on viral cores [16][56]. However, no HIV-1-induced ATF4 activation depending on these kinases has been reported yet.
To conclude, PERK and GCN2 may lead to ATF4 activation after HIV-1 infection, and the direct interaction of HIV-1 components with GCN2 and MARK2 may in turn control their ability to activate ATF4. The potential control of ATF4 by the new eIF2α kinases needs further investigation during HIV-1 infection.
2.2.2. Mitochondrial Stress Response, ATF4 and HIV-1
The contribution of HIV-1 infection to the induction of mitochondrial dysfunctions has been described earlier [57][58][59]. During HIV-1 infection, mitochondrial functions are compromised resulting in reduced oxidative phosphorylation (OXPHOS), ATP synthesis, gluconeogenesis, and β-oxidation. In addition to membrane depolarization and release of cytochrome C [60][61], HIV-1-induced alterations may also disrupt cellular homeostasis, increase oxidative stress, affect mitochondrial dynamics, and lead to the loss of mitochondrial DNA [60][62][63]. Mitochondrial dysfunctions are not only observed in CD4+ T cells, but also in myeloid cells such as neutrophils [64][65], monocytes [66], and CD8+ T cells [67], which are non-infected T cells. Therefore, indirect mechanisms may contribute to the alteration of mitochondrial functions during HIV-1 and SIV infections.
ATF4 is induced by several mitochondrial stress-like alterations affecting proteostasis, respiration, and mitochondrial membrane potential (MMP) loss [68][69][70]. A multi-omics study has suggested that ATF4 coordinates the mitochondrial stress response [70]. ATF4 induction was shown to depend on HRI in response to the respiratory chain and ATP synthesis disruptions [71][72]. Individual knockdown of each of the four original ISR kinases (i.e., HRI, PERK, PKR and GCN2) did not abolish the induction of ATF4 and its target genes. These results could suggest the role of either unknown or newly identified ISR kinases (as indicated above), or some redundancy.
In response to mitochondrial misfolded protein accumulation, ATF4 plays a significant role in cell homeostasis as a transcription factor of the canonical mitochondrial UPR (UPRmt) [69]. ATF4 is also associated with the transcription of genes encoding mitochondrial chaperonin and proteases [69]. For instance, Li et al. suggest that unfolded mitochondrial proteins would be degraded by lysosomes, leading to the increase in amino acids that would activate mTORC1 via lysosomal v-ATPase through a still-unknown mechanism during the UPRmt [73][74][75][76]. mTORC1 would then phosphorylate and activate ATF4, thereby triggering transcription of chaperone-encoding genes, and increasing the mitochondrial folding capacity [74].
ATF4 is also activated in response to alterations in mitochondrial dynamics. The deletion of optic atrophy protein 1 (OPA1), which is crucial for the fusion of inner membranes during mitochondrial fusion, and contributes to the release of apoptogenic factors [77][78], generates mitochondria-derived reactive oxygen species (ROS) and thus causes increased oxidative stress and death [62]. This process triggers ER stress and a PERK-dependent UPR, resulting in the transcriptional activation of ATF4 and other genes. This response initiates a catabolic program contributing to muscle loss and systemic aging [79]. The knockdown of Drp1, a major effector of mitochondria fission [80], leads to eIF2α phosphorylation and ATF4 activation in the liver [81]. Although modulation of OPA1 was not directly associated with ATF4 activation, it has been show that the OMA1 protease, which cleaves the mitochondrial protein DELE1, but also OPA1 [82][83], leads to the release in the cytosol of the DELE1′s carboxy-terminal domain that oligomerizes with HRI (Figure 1) [71][72][84].
2.2.3. The Viral Vpu Protein Stabilizes the ATF4 Protein
The HIV-1 viral protein U (Vpu) is an HIV-1 accessory protein that down-modulates CD4 and BST-2/tetherin. A cellular Skp, Cullin, F-Box (SCF) E3 ubiquitin ligase complex is recruited by Vpu to target CD4 for ubiquitination and proteasomal degradation [85]. This process involves the recruitment of the F-box β-transducin repeat-containing protein (βTrCP) [86][87][88][89]. Additionally, Vpu reduces BST-2/tetherin from the cell surface by preventing the trafficking of BST-2/tetherin to the plasma membrane from the trans Golgi network and/or the recycling endosome [90][91][92]. Furthermore, Vpu targets BST-2/tetherin for degradation, thus promoting viral progeny release and inhibiting NF-κB signaling [88][93].
The SCF-βTrCP E3 ubiquitin ligase complex has been reported to contribute in the degradation of ATF4 (Figure 1) [94]. Unlike its effect on CD4 and BST-2/tetherin, Vpu inhibits ATF4 βTrCP-mediated proteasomal degradation [95]. This apparent discrepancy in the effect of Vpu on βTrCP-dependent proteasomal degradation may be explained by the existence of two distinct paralogs of β-TrCP, βTrCP1/BTRC and βTrCP2/FBXW11 [96]. Recent work by Pickering et al. demonstrates that Vpu has contrasting effects on βTrCP1 and βTrCP2 and suggests that Vpu would induce proteasomal degradation mediated by βTrCP2 and inhibit βTrCP1-dependent protein degradation [97]. Thus, the contribution of viral proteins encoded by HIV-1 merits further investigation regarding the role of ATF4.
2.2.4. HIV-1 Antiretroviral Drugs Induce ATF4 Signaling
HIV antiretroviral therapy (ART) has drastically altered the course of HIV-1 infection, resulting in a major decrease in morbidity and mortality. However, drug side effects have been reported earlier, leading to their progressive replacements and the development of new molecules. Thus, mitochondrial damage was initially reported following the use of reverse transcriptase inhibitors (RTIs) and protease inhibitors (PIs) [98][99][100][101][102]. In addition to mitochondrial stress, it has been shown that Nelfinavir (PI) triggers an ATF4 transcriptional response associated with liver metabolic alterations that have been reported in PLWH [103] and causes cell cytotoxicity against ovarian cancer cells [104][105]. In addition to Nelfinavir, it has been shown that Lopinavir (PI) also increases the level of ATF4 transcript in SQ20B and FaDu cancer cell lines [106] but in a model-dependent manner because no effect was observed on the level of ATF4 transcript in a model of trophoblast cell differentiation [107]. While Zidovudine (AZT, RTI), which induces mitochondrial stress, was recently reported to extend the lifespan of C. elegans depending on ATF4 activation [108], a reduction in ATF4 expression was reported [109] in long-term Tenofovir disoproxil fumarate (TDF)-treated individuals presenting a decrease in bone mineral density. Therefore, the role of ATF4 remains to be clarified regarding the use of RTIs. Interestingly, an HIV-1 integrase inhibitor (IN), namely MK-2048, was shown to selectively kill HTLV-1–infected cells by inducing the PERK/ATF4/CHOP pathway [110]. This molecule could be also of interest for people living with HIV-1 (PLWH), in which triggering the death of viral infected cells may reduce the extent of viral reservoirs. Thus, several ARTs may provide a beneficial effect not only by tackling HIV viral replication but also by stimulating ER stress via ATF4 and thus facilitating the death of infected cells.
3. ATF4 Role during HIV-1 Replication
3.1. ATF4 Positively Regulates HIV-1 Cycle
The induction of ATF4 during HIV-1 infection raises the possibility that ATF4 may play a role in viral replication. Thus, it has been shown that the overexpression of ATF4 promotes viral replication, whereas its silencing suppresses HIV-1 replication [11][111]. Using compounds that inhibit GCN2 or PERK or that lead to increases in ATF4 levels, it has been shown that the induction of the ISR/ATF4 pathway reactivates HIV-1 in models of HIV-1 latency [12][13][112]. Altogether, these data strongly suggest a role for ATF4 in regulating HIV-1 replication both during the acute infection and the exit from latency. However, given the nature of infected cells that include memory CD4 T cells and T follicular helper cells (Tfh) [113][114][115][116][117], which represent the main reservoirs in visceral tissues, further analyses should be performed to elucidate the role of ATF4 in primary T cell subsets.
3.2. How ATF4 Favorizes HIV-1 Replication
3.2.1. ATF4 Binds to the HIV-1 LTR and Promotes Viral Gene Transcription
ATF4 can be a direct regulator of HIV-1 transcription. During HIV-1 infection, various cellular transcription factors including some bZIP domain proteins have been shown to bind the 5′ long terminal repeat (LTR)—that contains the transcriptional promoter of the viral genome of HIV-1—and regulates transcription of HIV-1 genes [118] (Figure 2).
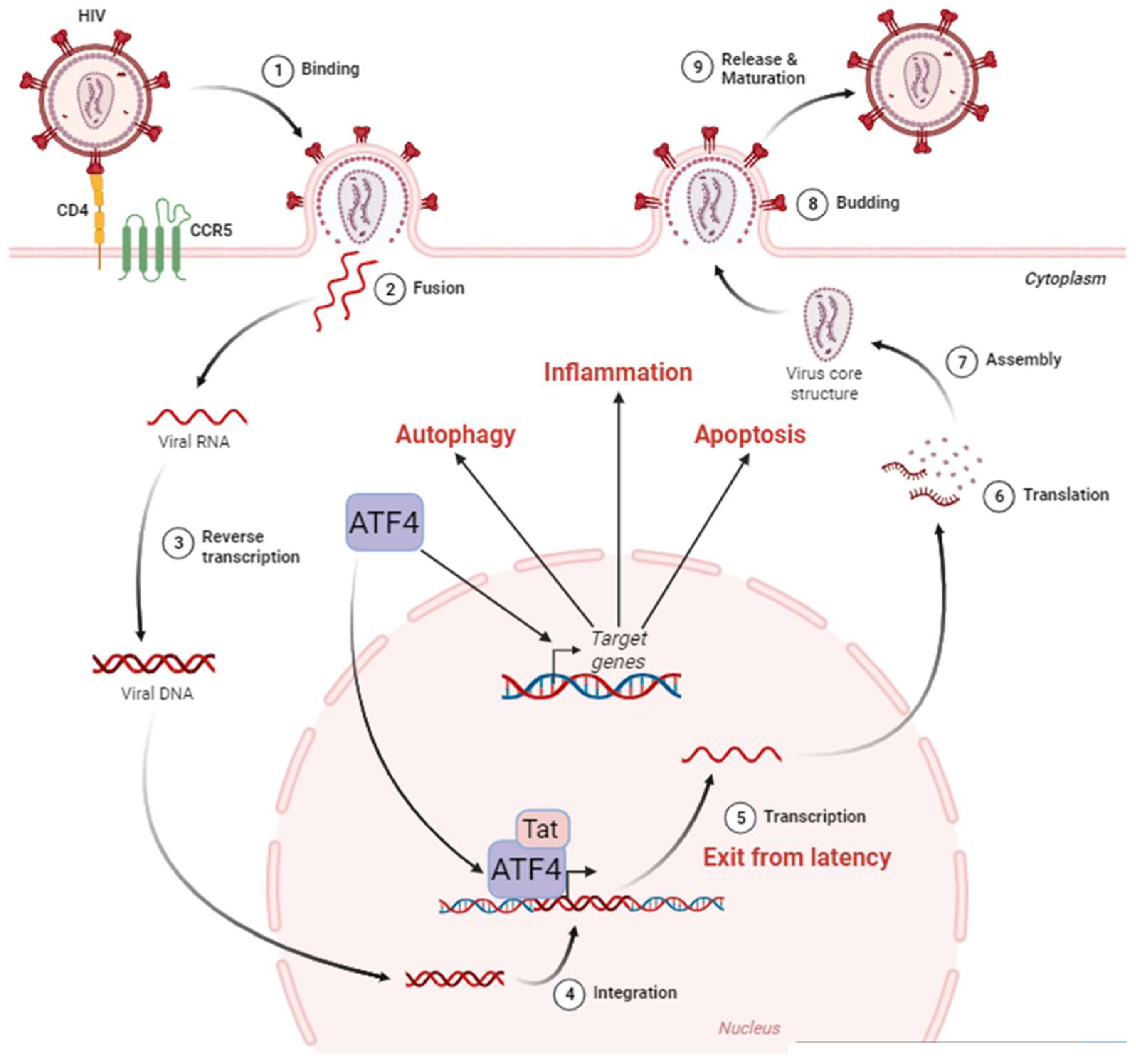
Figure 2. ATF4 in the HIV-1 cycle. The HIV-1 cycle begins with the attachment of the virus to the cell surface thanks to the CD4 and CCR5 receptors (1). This allows the fusion between the viral envelope and the host cell membrane (2). During this phase, the viral nucleocapsid enters the cell cytoplasm. Single-stranded RNA molecules are then retro-transcribed in the nucleus (3). The viral DNA then integrates into the host cell genome (4), where it can remain latent until reactivation. The exit from the latency is stimulated by ATF4, which, in cooperation with the viral protein Tat, activates the HIV-1 promoter located in the LTR region of the viral DNA leading to viral RNA (5). These HIV-1 messenger RNAs are exported from the nucleus to be translated (6) into proteins that assemble to form the internal structure of viral particles (7). Budding from the cell membrane (8) culminates with the release and maturation of viral particles in the extracellular environment (9). ATF4 is also capable of inducing the activation of target genes involved in cellular processes such as autophagy, inflammation, and apoptosis. Adapted from “Disease Mechanism–Infectious Diseases, HIV Replication Cycle”, by BioRender.com. Retrieved from https://app.biorender.com/biorender-templates (accessed on 18 January 2024).
ATF4 induces HIV-1’s gene transcription and regulates the Human T-cell Leukemia Virus (HTLV) promoter [119]. As an ATF/CREB family member, ATF4 binds to the C/EBP-ATF consensus sequence (TGACGT (C/A) (G/A)). Two C/EBP-ATF-binding sites were first identified in the LTR of HIV-1, but super-electrophoretic mobility shift assay (EMSAs) failed to show the binding of ATF4 to these regions [120][121]. A bioinformatics approach led by Jiang et al. identified additional potential C/EBP-ATF-binding sites in the LTR [12]. Caselli et al. reported that co-expression of Tat and ATF4 induced higher LTR transcription than the sole expression of Tat, suggesting a synergistic interaction between the two proteins [11]. ChIP experiments showed that ATF4 binds to the LTR sequence. This interaction is positively regulated by Forkhead box O 1 (FOXO1) inhibition and amino acids deprival [12][112]. Further works suggest that Tat may be unnecessary for ATF4-mediated HIV-1 promoter activation [11][111]. However, this effect has only been observed in HeLa cells, but not in Jurkat and 293 T cells [11].
ATF4 can also bind to sequences that differ from the C/EBP-ATF consensus sequence, depending on its dimerization partners (Figure 3) [1][122][123]. ATF4 can dimerize with Jun and Fos [124] that heterodimerize to form the AP-1 transcription factor [124]. The fact that the HIV-1 promoter presents an AP-1-binding site could suggest that a heterodimer of ATF4 with Jun or Fos can be formed to activate the LTR activity. However, the partners able to dimerize with ATF4 to control HIV-1 transcription are still unknown.
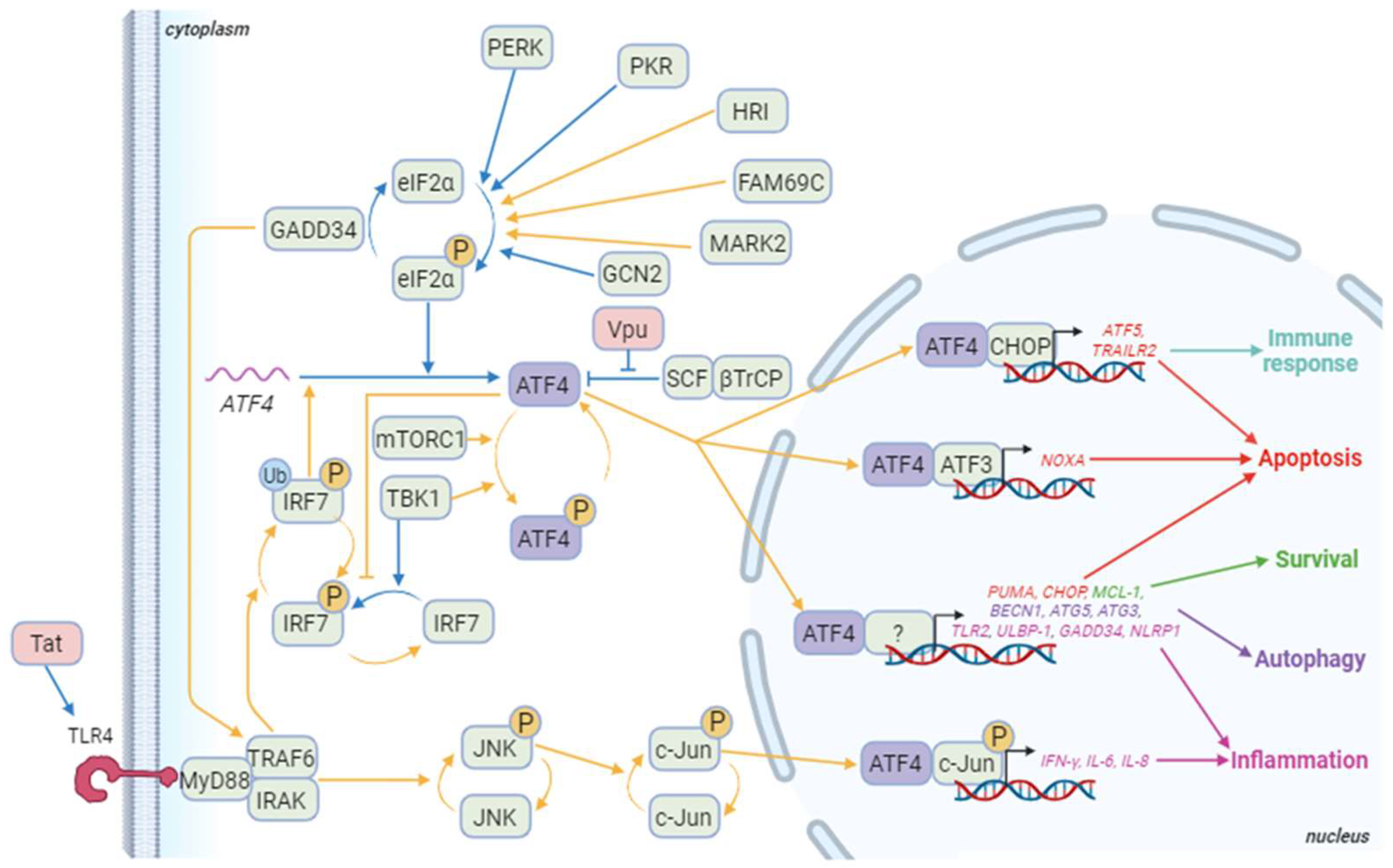
Figure 3. ATF4 signaling pathways during HIV-1 infection. Blue arrows correspond to interactions demonstrated in HIV-1 literature. Yellow arrows correspond to other contexts. Besides ATF4, cellular proteins appear in green and HIV-1 proteins in red. The genes in red are some of the pro-apoptotic genes induced by ATF4/ATF3 or ATF4/CHOP dimers and ATF4 dimerizing with a partner that remains to be determined. ATF5 also regulates the immune response at least by controlling immune cell differentiation. ATF4 also controls the expression of several pro-apoptotic genes including TID1/DNAJA3, G0S2, and TP53BP2. The genes in green are involved in ATF4-mediated survival as TMBIM5. Genes in purple are implicated in ATF4-induced autophagy like WIPI1, ATG12, ATG10, ATG16L1, ATG7, MAP1lc3B, GABARAPl2, p62/SQSTM1, NBR1, and REDD1. Finally, genes in pink are genes related to inflammation that are induced by ATF4 in a dimer with phosphorylated c-Jun or an undefined partner. Other genes activated by the phosphorylated ATF4/c-Jun dimer include RANTES and sICAM-1. Not shown in this diagram is IRF7 activation, which induces the production of type I and II interferons, and then activates the PKR/eIF2α/ATF4 pathway. Created with BioRender.com (accessed on the 18 January 2024).
Five to twenty percent of HIV-1-infected patients are co-infected by the Hepatitis B virus (HBV). The X protein of HBV has been shown to regulate HIV-1 transcription by stimulating the binding of ATF4 to the LTR of HIV-1 [125]. ATF4 can also directly interact with the HTLV-1 transcriptional activator Tax protein and act as an adapter between Tax and the HTLV-1 promoter [119][126]. Therefore, co-infections may provide a permissive environment for ATF4 to activate the HIV-1 promoter.
3.2.2. ATF4, HIV-1 and Apoptosis
It is well known that HIV-1 pathogenicity is associated with the depletion of CD4+ T cells leading to the development of AIDS. In this context, it has been shown that a form of programmed cell death, namely apoptosis, may contribute to the depletion of CD4+ T cells and be associated with pathogenicity [6][127][128]. Both in vitro and in vivo, HIV-1 and SIV infections mediate T cell death [8][129] through the intrinsic apoptotic pathway that is characterized by the expression of the pro-apoptotic proteins Bax, Bak, and Bim [130], leading to the release of apoptogenic mitochondrial factors and consequently to caspase activation. CD4+ T cell apoptosis also involves the extrinsic pathway in which Fas (CD95) is critical [131][132][133][134]. Thus, caspase inhibition prevents in vivo the depletion of CD4+ T cells and delays the progression to AIDS [7].
It has been shown that Tat up-regulates UPR mediators with an increase in the transcript levels of ATF4, CHOP and the pro-apoptotic BH3-only BIM (BCL2L11), thus leading to apoptosis [14]. Of interest, ATF4 mediates cell death through the activation of the bZIP transcription factor CHOP [3], which also induces the transcription of BIM and of the pro-apoptotic BH3-only PUMA (also known as BBC3) [135][136]. Additionally, the CHOP-ATF4 dimer up-regulates ATF5, NOXA (also known as PMAIP1), APAF-1, and TXNIP, which in turn amplify cell death [137][138][139]. ATF4 also promotes degradation of the caspase inhibitor X chromosome-linked inhibitor of apoptosis (XIAP) protein through the ubiquitin-proteasome system, which allows the activation of caspases [140].
Altogether, the identified ATF4 targets represent multiple new mechanisms to be explored by which ATF4 could regulate cell death or survival of HIV-1-infected cells.
3.2.3. ATF4, HIV-1 and Autophagy
Autophagy is a pro-survival intracellular catabolic process that targets protein aggregates and damaged organelles in response to stress. ATF4 activates the transcription of genes involved in the initiation (BECN1), in the elongation of the phagophore and the maturation of the autophagosome (WIPI1, ATG12, ATG5, ATG10, ATG16L1, ATG3, ATG7, MAP1lc3B, GABARAPL2), and in the selective clearance of cargo (p62/SQSTM1 and NBR1) [122][141]. Interestingly, ATF4 triggers the transcriptional activation of the autophagy genes directly as a homo or heterodimer with CHOP or indirectly through CHOP induction (Figure 3) [141]. The ratio between ATF4 and CHOP is decisive for the regulation of these genes, underpinning a fine-tuned control of the different stages of autophagy, the subtleties of which remain to be elucidated. ATF4 can also act upstream of autophagy to induce this process; ATF4 can increase the expression of the gene encoding Regulated in development and DNA damage response 1 (REDD1; also known as Ddit4), which suppresses mTOR complex 1 (mTORC1) activity, allowing autophagy induction upon ER stress and starvation [142][143][144].
HIV-1 modulates autophagy in a cell-type-dependent manner [145][146]. The Env protein found at the cell surface of infected cells induces an autophagy-dependent cell death of bystander CD4+ T cells [147]. Thus, ARF6, which promotes autophagosome biogenesis by facilitating endocytic uptake of the plasma membrane into autophagosome precursors, has been proposed to promote the fusion of HIV-1 envelope with the plasma membrane, therefore permitting the entry of HIV-1 in CD4+ T lymphocytes [148][149]. Other groups have reported that HIV-1 inhibits autophagy [150][151]. After virus entry, the Vpr protein decreases the amount of proteins like MAP1LC3 and BECN1 [152]. Furthermore, although autophagy is initiated in primary CD4+ T cells infected with HIV-1, this process aborts due to a lysosome destabilization by DRAM, a p53-inducible gene. This permeabilization of lysosomes and resulting cell death has been proposed as an altruistic self-defense mechanism limiting viral dissemination by the elimination of infected cells [153]. Despite the obvious link between ATF4 and autophagy, the role of ATF4 in autophagy in the context of HIV-1 infection remains poorly documented.
3.2.4. Immune Response and ATF4 Activation during HIV-1 Infection
ATF4, innate immunity and HIV-1
Pattern recognition receptors (PRRs) are fundamental molecules for innate immune response recognizing pathogen-associated molecular patterns (PAMPs). PRRs bind to PAMPs and trigger cell signaling cascades. PRRs include Toll-like receptors (TLRs), Nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), RIG-I-like receptors (RLRs), and C-type lectin receptors (CLRs). For example, TLR7 and TLR8 can recognize single-stranded RNAs, which are present in the HIV-1 genome [154], leading to Interferon regulatory factor 3 and 7 (IRF3 and IRF7) activations and inducing robust expression of type I interferon (IFN) genes [155]. ATF4 inhibits IRF7 transcription but also its activation by inhibiting TANK-binding kinase 1 (TBK1) and IκB kinase (IKK)ε-mediated IRF7 phosphorylation [156]. To be activated, IRF7 also requires its ubiquitination by Tumor necrosis factor receptor-associated factor 6 (TRAF6) (Figure 3). GADD34 that binds to and inhibits TRAF6 activity is an ATF4 target [157]. Its transcript levels have been shown to be increased in HIV-1-infected cells in a p53-dependent manner [158]. Inhibition of TRAF6 enhanced HIV-1 replication in macrophages [159], whereas GADD34 reduced viral protein expression [160]. Therefore, the role of ATF4 and its targets merits to be further explored in innate immunity.
ATF4, cellular immunity and HIV-1
Viruses have developed a diverse array of strategies to manipulate host cell metabolism and reorientate metabolic resources toward their benefit [63]. Current knowledge on T-cell metabolism in HIV-1 infection suggests that HIV-1 takes advantage of the glycolytic process in CD4+ T cells to infect them and to boost viral replication [161][162][163]. It has been shown that IL-7-mediated thymocytes survival through the increase in Glut1 protein levels at the cell surface, leading to glucose uptake and favoring viral infection [164]. Interestingly, ATF4 can influence a network of genes responsible for metabolic flux when activated by an extracellular oxidizing environment [165]. Thus, ATF4 up-regulates genes involved in glycolysis and promotes the anaplerotic flux through enhanced glutaminolysis, which plays a role in T cell growth in oxygen- or amino-acid-deprived environments [165]. Glutaminolysis fuels the tricarboxylic acid (TCA) cycle and oxidative phosphorylation (OXPHOS) in T-cell receptor-stimulated naïve CD4 subsets, as well as memory CD4 subsets [166]. This balance in the bioenergetics pathways is essential for T cell polarization [167]. ATF4 has also been implicated in regulating the differentiation of naïve T cells in T helper 17 cells (Th17). T cells from ATF4-deficient mice are characterized by a diminished Th1 differentiation and an elevation in factors promoting Th17 commutation, such as Interleukin-17 (IL-17) [165]. This association is further supported by the effect of the small molecule halofuginone (HF), which inhibits Th17 differentiation in both mice and humans and increases ATF4 protein levels [168]. Notably, adding amino acids in excess counteracts the inhibition of Th17 differentiation by HF, suggesting the involvement of the amino acid starvation response mediated by ATF4 in targeting Th17 differentiation. Interestingly, Th17 cells are highly vulnerable to HIV-1 infection [169], and this population represents a major viral reservoir under ART [170]. The loss of Th17 homeostasis contributes to disease progression in SIV-infected rhesus macaques associated with the loss of intestinal epithelial integrity, leading to microbial translocation [171][172][173][174]. This alteration in the balance of T cell subsets persists despite ART [175]. Given the role of ATF4 in regulating mitochondrial and reticulum stress-associated apoptosis, a contribution of ATF4 dysregulation in the alteration of the Th17 balance at the mucosal barrier cannot be excluded. Taken together, these results suggest that the induction of ATF4 observed in CD4+ T lymphocytes during HIV-1 infection [13] may contribute to the increased production of energy and amino acids required for viral replication.
References
- Ameri, K.; Harris, A.L. Activating Transcription Factor 4. Int. J. Biochem. Cell Biol. 2008, 40, 14–21.
- Kasai, S.; Yamazaki, H.; Tanji, K.; Engler, M.J.; Matsumiya, T.; Itoh, K. Role of the ISR-ATF4 Pathway and Its Cross Talk with Nrf2 in Mitochondrial Quality Control. J. Clin. Biochem. Nutr. 2019, 64, 1–12.
- Pakos-Zebrucka, K.; Koryga, I.; Mnich, K.; Ljujic, M.; Samali, A.; Gorman, A.M. The Integrated Stress Response. EMBO Rep. 2016, 17, 1374–1395.
- Angelastro, J.M. Targeting ATF5 in Cancer. Trends Cancer 2017, 3, 471–474.
- Greene, L.A.; Lee, H.Y.; Angelastro, J.M. The Transcription Factor ATF5: Role in Neurodevelopment and Neural Tumors. J. Neurochem. 2009, 108, 11–22.
- Estaquier, J.; Idziorek, T.; de Bels, F.; Barré-Sinoussi, F.; Hurtrel, B.; Aubertin, A.M.; Venet, A.; Mehtali, M.; Muchmore, E.; Michel, P.; et al. Programmed Cell Death and AIDS: Significance of T-Cell Apoptosis in Pathogenic and Nonpathogenic Primate Lentiviral Infections. Proc. Natl. Acad. Sci. USA 1994, 91, 9431–9435.
- Laforge, M.; Silvestre, R.; Rodrigues, V.; Garibal, J.; Campillo-Gimenez, L.; Mouhamad, S.; Monceaux, V.; Cumont, M.-C.; Rabezanahary, H.; Pruvost, A.; et al. The Anti-Caspase Inhibitor Q-VD-OPH Prevents AIDS Disease Progression in SIV-Infected Rhesus Macaques. J. Clin. Investig. 2018, 128, 1627–1640.
- Monceaux, V.; Estaquier, J.; Février, M.; Cumont, M.-C.; Rivière, Y.; Aubertin, A.-M.; Ameisen, J.C.; Hurtrel, B. Extensive Apoptosis in Lymphoid Organs during Primary SIV Infection Predicts Rapid Progression towards AIDS. AIDS 2003, 17, 1585–1596.
- Cumont, M.-C.; Diop, O.; Vaslin, B.; Elbim, C.; Viollet, L.; Monceaux, V.; Lay, S.; Silvestri, G.; Le Grand, R.; Müller-Trutwin, M.; et al. Early Divergence in Lymphoid Tissue Apoptosis between Pathogenic and Nonpathogenic Simian Immunodeficiency Virus Infections of Nonhuman Primates. J. Virol. 2008, 82, 1175–1184.
- Viollet, L.; Monceaux, V.; Petit, F.; Ho Tsong Fang, R.; Cumont, M.-C.; Hurtrel, B.; Estaquier, J. Death of CD4+ T Cells from Lymph Nodes during Primary SIVmac251 Infection Predicts the Rate of AIDS Progression. J. Immunol. 2006, 177, 6685–6694.
- Caselli, E.; Benedetti, S.; Gentili, V.; Grigolato, J.; Di Luca, D. Short Communication: Activating Transcription Factor 4 (ATF4) Promotes HIV Type 1 Activation. AIDS Res. Hum. Retroviruses 2012, 28, 907–912.
- Jiang, G.; Santos Rocha, C.; Hirao, L.A.; Mendes, E.A.; Tang, Y.; Thompson, G.R.; Wong, J.K.; Dandekar, S. HIV Exploits Antiviral Host Innate GCN2-ATF4 Signaling for Establishing Viral Replication Early in Infection. mBio 2017, 8, e01518-16.
- Li, D.; Wong, L.M.; Tang, Y.; Allard, B.; James, K.S.; Thompson, G.R.; Dandekar, S.; Browne, E.P.; Li, Q.; Simon, J.M.; et al. Depletion of HIV Reservoir by Activation of ISR Signaling in Resting CD4+T Cells. iScience 2023, 26, 105743.
- Campestrini, J.; Silveira, D.B.; Pinto, A.R. HIV-1 Tat-Induced Bystander Apoptosis in Jurkat Cells Involves Unfolded Protein Responses. Cell Biochem. Funct. 2018, 36, 377–386.
- Lu, Y.-N.; Kavianpour, S.; Zhang, T.; Zhang, X.; Nguyen, D.; Thombre, R.; He, L.; Wang, J. MARK2 Phosphorylates eIF2α in Response to Proteotoxic Stress. PLoS Biol. 2021, 19, e3001096.
- Wu, Z.; Mei, F.; Gan, Y.; Liu, A.; Hu, J.; Jin, Y.; Yin, Y. FAM69C Functions as a Kinase for eIF2α and Promotes Stress Granule Assembly. EMBO Rep. 2023, 24, e55641.
- Liu, Y.; Wang, M.; Cheng, A.; Yang, Q.; Wu, Y.; Jia, R.; Liu, M.; Zhu, D.; Chen, S.; Zhang, S.; et al. The Role of Host eIF2α in Viral Infection. Virol. J. 2020, 17, 112.
- Galabru, J.; Hovanessian, A. Autophosphorylation of the Protein Kinase Dependent on Double-Stranded RNA. J. Biol. Chem. 1987, 262, 15538–15544.
- García, M.A.; Meurs, E.F.; Esteban, M. The dsRNA Protein Kinase PKR: Virus and Cell Control. Biochimie 2007, 89, 799–811.
- Carpick, B.W.; Graziano, V.; Schneider, D.; Maitra, R.K.; Lee, X.; Williams, B.R. Characterization of the Solution Complex between the Interferon-Induced, Double-Stranded RNA-Activated Protein Kinase and HIV-I Trans-Activating Region RNA. J. Biol. Chem. 1997, 272, 9510–9516.
- Kim, I.; Liu, C.W.; Puglisi, J.D. Specific Recognition of HIV TAR RNA by the dsRNA Binding Domains (dsRBD1-dsRBD2) of PKR. J. Mol. Biol. 2006, 358, 430–442.
- Maitra, R.K.; McMillan, N.A.; Desai, S.; McSwiggen, J.; Hovanessian, A.G.; Sen, G.; Williams, B.R.; Silverman, R.H. HIV-1 TAR RNA Has an Intrinsic Ability to Activate Interferon-Inducible Enzymes. Virology 1994, 204, 823–827.
- Spanggord, R.J.; Vuyisich, M.; Beal, P.A. Identification of Binding Sites for Both dsRBMs of PKR on Kinase-Activating and Kinase-Inhibiting RNA Ligands. Biochemistry 2002, 41, 4511–4520.
- Clerzius, G.; Gélinas, J.-F.; Gatignol, A. Multiple Levels of PKR Inhibition during HIV-1 Replication. Rev. Med. Virol. 2011, 21, 42–53.
- Lee, E.-S.; Yoon, C.-H.; Kim, Y.-S.; Bae, Y.-S. The Double-Strand RNA-Dependent Protein Kinase PKR Plays a Significant Role in a Sustained ER Stress-Induced Apoptosis. FEBS Lett. 2007, 581, 4325–4332.
- Benkirane, M.; Neuveut, C.; Chun, R.F.; Smith, S.M.; Samuel, C.E.; Gatignol, A.; Jeang, K.T. Oncogenic Potential of TAR RNA Binding Protein TRBP and Its Regulatory Interaction with RNA-Dependent Protein Kinase PKR. EMBO J. 1997, 16, 611–624.
- Blair, E.D.; Roberts, C.M.; Snowden, B.W.; Gatignol, A.; Benkirane, M.; Jeang, K.-T. Expression of TAR RNA-Binding Protein in Baculovirus and Co-Immunoprecipitation with Insect Cell Protein Kinase. J. Biomed. Sci. 1995, 2, 322–329.
- Clerzius, G.; Gélinas, J.-F.; Daher, A.; Bonnet, M.; Meurs, E.F.; Gatignol, A. ADAR1 Interacts with PKR during Human Immunodeficiency Virus Infection of Lymphocytes and Contributes to Viral Replication. J. Virol. 2009, 83, 10119–10128.
- Cole, J.L. Activation of PKR: An Open and Shut Case? Trends Biochem. Sci. 2007, 32, 57–62.
- Daher, A.; Longuet, M.; Dorin, D.; Bois, F.; Segeral, E.; Bannwarth, S.; Battisti, P.L.; Purcell, D.F.; Benarous, R.; Vaquero, C.; et al. Two Dimerization Domains in the Trans-Activation Response RNA-Binding Protein (TRBP) Individually Reverse the Protein Kinase R Inhibition of HIV-1 Long Terminal Repeat Expression. J. Biol. Chem. 2001, 276, 33899–33905.
- Duarte, M.; Graham, K.; Daher, A.; Battisti, P.L.; Bannwarth, S.; Segeral, E.; Jeang, K.T.; Gatignol, A. Characterization of TRBP1 and TRBP2. Stable Stem-Loop Structure at the 5′ End of TRBP2 mRNA Resembles HIV-1 TAR and Is Not Found in Its Processed Pseudogene. J. Biomed. Sci. 2000, 7, 494–506.
- Lemaire, P.A.; Anderson, E.; Lary, J.; Cole, J.L. Mechanism of PKR Activation by dsRNA. J. Mol. Biol. 2008, 381, 351–360.
- Brand, S.R.; Kobayashi, R.; Mathews, M.B. The Tat Protein of Human Immunodeficiency Virus Type 1 Is a Substrate and Inhibitor of the Interferon-Induced, Virally Activated Protein Kinase, PKR. J. Biol. Chem. 1997, 272, 8388–8395.
- Cai, R.; Carpick, B.; Chun, R.F.; Jeang, K.T.; Williams, B.R. HIV-I TAT Inhibits PKR Activity by Both RNA-Dependent and RNA-Independent Mechanisms. Arch. Biochem. Biophys. 2000, 373, 361–367.
- McMillan, N.A.; Chun, R.F.; Siderovski, D.P.; Galabru, J.; Toone, W.M.; Samuel, C.E.; Mak, T.W.; Hovanessian, A.G.; Jeang, K.T.; Williams, B.R. HIV-1 Tat Directly Interacts with the Interferon-Induced, Double-Stranded RNA-Dependent Kinase, PKR. Virology 1995, 213, 413–424.
- Mehrbod, P.; Ande, S.R.; Alizadeh, J.; Rahimizadeh, S.; Shariati, A.; Malek, H.; Hashemi, M.; Glover, K.K.M.; Sher, A.A.; Coombs, K.M.; et al. The Roles of Apoptosis, Autophagy and Unfolded Protein Response in Arbovirus, Influenza Virus, and HIV Infections. Virulence 2019, 10, 376–413.
- Ma, R.; Yang, L.; Niu, F.; Buch, S. HIV Tat-Mediated Induction of Human Brain Microvascular Endothelial Cell Apoptosis Involves Endoplasmic Reticulum Stress and Mitochondrial Dysfunction. Mol. Neurobiol. 2016, 53, 132–142.
- Tripathi, A.; Iyer, K.; Mitra, D. HIV-1 Replication Requires Optimal Activation of the Unfolded Protein Response. FEBS Lett. 2023, 597, 2908–2930.
- Hortin, G.L.; Landt, M.; Powderly, W.G. Changes in Plasma Amino Acid Concentrations in Response to HIV-1 Infection. Clin. Chem. 1994, 40, 785–789.
- Dröge, W.; Murthy, K.K.; Stahl-Hennig, C.; Hartung, S.; Plesker, R.; Rouse, S.; Peterhans, E.; Kinscherf, R.; Fischbach, T.; Eck, H.P. Plasma Amino Acid Dysregulation after Lentiviral Infection. AIDS Res. Hum. Retroviruses 1993, 9, 807–809.
- Mody, A.; Bartz, S.; Hornik, C.P.; Kiyimba, T.; Bain, J.; Muehlbauer, M.; Kiboneka, E.; Stevens, R.; St Peter, J.V.; Newgard, C.B.; et al. Effects of HIV Infection on the Metabolic and Hormonal Status of Children with Severe Acute Malnutrition. PLoS ONE 2014, 9, e102233.
- Stone, J.D.; Heise, C.C.; Miller, C.J.; Halsted, C.H.; Dandekar, S. Development of Malabsorption and Nutritional Complications in Simian Immunodeficiency Virus-Infected Rhesus Macaques. AIDS 1994, 8, 1245–1256.
- Cosnefroy, O.; Jaspart, A.; Calmels, C.; Parissi, V.; Fleury, H.; Ventura, M.; Reigadas, S.; Andréola, M.-L. Activation of GCN2 upon HIV-1 Infection and Inhibition of Translation. Cell Mol. Life Sci. 2013, 70, 2411–2421.
- Jaspart, A.; Calmels, C.; Cosnefroy, O.; Bellecave, P.; Pinson, P.; Claverol, S.; Guyonnet-Dupérat, V.; Dartigues, B.; Benleulmi, M.S.; Mauro, E.; et al. GCN2 Phosphorylates HIV-1 Integrase and Decreases HIV-1 Replication by Limiting Viral Integration. Sci. Rep. 2017, 7, 2283.
- del Pino, J.; Jiménez, J.L.; Ventoso, I.; Castelló, A.; Muñoz-Fernández, M.Á.; de Haro, C.; Berlanga, J.J. GCN2 Has Inhibitory Effect on Human Immunodeficiency Virus-1 Protein Synthesis and Is Cleaved upon Viral Infection. PLoS ONE 2012, 7, e47272.
- Lutz, M.M.; Worth, M.P.; Hinchman, M.M.; Parker, J.S.L.; Ledgerwood, E.D. Mammalian Orthoreovirus Infection Is Enhanced in Cells Pre-Treated with Sodium Arsenite. Viruses 2019, 11, 563.
- Lee, J.; Stone, J.; Desai, P.; Kosowicz, J.G.; Liu, J.O.; Ambinder, R.F. Arsenicals, the Integrated Stress Response, and Epstein-Barr Virus Lytic Gene Expression. Viruses 2021, 13, 812.
- McEwen, E.; Kedersha, N.; Song, B.; Scheuner, D.; Gilks, N.; Han, A.; Chen, J.-J.; Anderson, P.; Kaufman, R.J. Heme-Regulated Inhibitor Kinase-Mediated Phosphorylation of Eukaryotic Translation Initiation Factor 2 Inhibits Translation, Induces Stress Granule Formation, and Mediates Survival upon Arsenite Exposure. J. Biol. Chem. 2005, 280, 16925–16933.
- Mukherjee, T.; Ramaglia, V.; Abdel-Nour, M.; Bianchi, A.A.; Tsalikis, J.; Chau, H.N.; Kalia, S.K.; Kalia, L.V.; Chen, J.-J.; Arnoult, D.; et al. The eIF2α Kinase HRI Triggers the Autophagic Clearance of Cytosolic Protein Aggregates. J. Biol. Chem. 2021, 296, 100050.
- Huang, P.; Peslak, S.A.; Lan, X.; Khandros, E.; Yano, J.A.; Sharma, M.; Keller, C.A.; Giardine, B.; Qin, K.; Abdulmalik, O.; et al. The HRI-Regulated Transcription Factor ATF4 Activates BCL11A Transcription to Silence Fetal Hemoglobin Expression. Blood 2020, 135, 2121–2132.
- Suragani, R.N.V.S.; Zachariah, R.S.; Velazquez, J.G.; Liu, S.; Sun, C.-W.; Townes, T.M.; Chen, J.-J. Heme-Regulated eIF2α Kinase Activated Atf4 Signaling Pathway in Oxidative Stress and Erythropoiesis. Blood 2012, 119, 5276–5284.
- Protzer, U.; Seyfried, S.; Quasdorff, M.; Sass, G.; Svorcova, M.; Webb, D.; Bohne, F.; Hösel, M.; Schirmacher, P.; Tiegs, G. Antiviral Activity and Hepatoprotection by Heme Oxygenase-1 in Hepatitis B Virus Infection. Gastroenterology 2007, 133, 1156–1165.
- Zhu, Z.; Wilson, A.T.; Mathahs, M.M.; Wen, F.; Brown, K.E.; Luxon, B.A.; Schmidt, W.N. Heme Oxygenase-1 Suppresses Hepatitis C Virus Replication and Increases Resistance of Hepatocytes to Oxidant Injury. Hepatology 2008, 48, 1430–1439.
- Devadas, K.; Dhawan, S. Hemin Activation Ameliorates HIV-1 Infection via Heme Oxygenase-1 Induction. J. Immunol. 2006, 176, 4252–4257.
- Ambegaokar, S.S.; Kolson, D.L. Heme Oxygenase-1 Dysregulation in the Brain: Implications for HIV-Associated Neurocognitive Disorders. Curr. HIV Res. 2014, 12, 174–188.
- Malikov, V.; Naghavi, M.H. Localized Phosphorylation of a Kinesin-1 Adaptor by a Capsid-Associated Kinase Regulates HIV-1 Motility and Uncoating. Cell Rep. 2017, 20, 2792–2799.
- Cossarizza, A.; Mussini, C.; Mongiardo, N.; Borghi, V.; Sabbatini, A.; De Rienzo, B.; Franceschi, C. Mitochondria Alterations and Dramatic Tendency to Undergo Apoptosis in Peripheral Blood Lymphocytes during Acute HIV Syndrome. AIDS 1997, 11, 19–26.
- Macho, A.; Castedo, M.; Marchetti, P.; Aguilar, J.J.; Decaudin, D.; Zamzami, N.; Girard, P.M.; Uriel, J.; Kroemer, G. Mitochondrial Dysfunctions in Circulating T Lymphocytes from Human Immunodeficiency Virus-1 Carriers. Blood 1995, 86, 2481–2487.
- Petit, F.; Arnoult, D.; Lelièvre, J.-D.; Moutouh-de Parseval, L.; Hance, A.J.; Schneider, P.; Corbeil, J.; Ameisen, J.C.; Estaquier, J. Productive HIV-1 Infection of Primary CD4+ T Cells Induces Mitochondrial Membrane Permeabilization Leading to a Caspase-Independent Cell Death. J. Biol. Chem. 2002, 277, 1477–1487.
- Arnoult, D.; Petit, F.; Lelièvre, J.-D.; Estaquier, J. Mitochondria in HIV-1-Induced Apoptosis. Biochem. Biophys. Res. Commun. 2003, 304, 561–574.
- Arnoult, D.; Petit, F.; Lelièvre, J.D.; Lecossier, D.; Hance, A.; Monceaux, V.; Hurtrel, B.; Ho Tsong Fang, R.; Ameisen, J.C.; Estaquier, J. Caspase-Dependent and-Independent T-Cell Death Pathways in Pathogenic Simian Immunodeficiency Virus Infection: Relationship to Disease Progression. Cell Death Differ. 2003, 10, 1240–1252.
- Estaquier, J.; Vallette, F.; Vayssiere, J.-L.; Mignotte, B. The Mitochondrial Pathways of Apoptosis. Adv. Exp. Med. Biol. 2012, 942, 157–183.
- Mesquita, I.; Estaquier, J. Viral Manipulation of the Host Metabolic Network. Exp. Suppl. 2018, 109, 377–401.
- Elbim, C.; Monceaux, V.; François, S.; Hurtrel, B.; Gougerot-Pocidalo, M.-A.; Estaquier, J. Increased Neutrophil Apoptosis in Chronically SIV-Infected Macaques. Retrovirology 2009, 6, 29.
- Elbim, C.; Monceaux, V.; Mueller, Y.M.; Lewis, M.G.; François, S.; Diop, O.; Akarid, K.; Hurtrel, B.; Gougerot-Pocidalo, M.-A.; Lévy, Y.; et al. Early Divergence in Neutrophil Apoptosis between Pathogenic and Nonpathogenic Simian Immunodeficiency Virus Infections of Nonhuman Primates. J. Immunol. 2008, 181, 8613–8623.
- Elbim, C.; Pillet, S.; Prevost, M.H.; Preira, A.; Girard, P.M.; Rogine, N.; Matusani, H.; Hakim, J.; Israel, N.; Gougerot-Pocidalo, M.A. Redox and Activation Status of Monocytes from Human Immunodeficiency Virus-Infected Patients: Relationship with Viral Load. J. Virol. 1999, 73, 4561–4566.
- Cumont, M.C.; Monceaux, V.; Viollet, L.; Lay, S.; Parker, R.; Hurtrel, B.; Estaquier, J. TGF-Beta in Intestinal Lymphoid Organs Contributes to the Death of Armed Effector CD8 T Cells and Is Associated with the Absence of Virus Containment in Rhesus Macaques Infected with the Simian Immunodeficiency Virus. Cell Death Differ. 2007, 14, 1747–1758.
- Kasai, S.; Yasumoto, K.-I.; Sogawa, K. Attenuation of Inhibitory PAS Domain Protein-Induced Cell Death by Synthetic Peptides Derived from Mcl-1 Transmenbrane Domain. Cell Death Discov. 2021, 7, 92.
- Münch, C. The Different Axes of the Mammalian Mitochondrial Unfolded Protein Response. BMC Biol. 2018, 16, 81.
- Quirós, P.M.; Prado, M.A.; Zamboni, N.; D’Amico, D.; Williams, R.W.; Finley, D.; Gygi, S.P.; Auwerx, J. Multi-Omics Analysis Identifies ATF4 as a Key Regulator of the Mitochondrial Stress Response in Mammals. J. Cell Biol. 2017, 216, 2027–2045.
- Fessler, E.; Eckl, E.-M.; Schmitt, S.; Mancilla, I.A.; Meyer-Bender, M.F.; Hanf, M.; Philippou-Massier, J.; Krebs, S.; Zischka, H.; Jae, L.T. A Pathway Coordinated by DELE1 Relays Mitochondrial Stress to the Cytosol. Nature 2020, 579, 433–437.
- Guo, X.; Aviles, G.; Liu, Y.; Tian, R.; Unger, B.A.; Lin, Y.-H.T.; Wiita, A.P.; Xu, K.; Correia, M.A.; Kampmann, M. Mitochondrial Stress Is Relayed to the Cytosol by an OMA1-DELE1-HRI Pathway. Nature 2020, 579, 427–432.
- Lawrence, R.E.; Zoncu, R. The Lysosome as a Cellular Centre for Signalling, Metabolism and Quality Control. Nat. Cell Biol. 2019, 21, 133–142.
- Li, T.Y.; Wang, Q.; Gao, A.W.; Li, X.; Sun, Y.; Mottis, A.; Shong, M.; Auwerx, J. Lysosomes Mediate the Mitochondrial UPR via mTORC1-Dependent ATF4 Phosphorylation. Cell Discov. 2023, 9, 92.
- Wolfson, R.L.; Sabatini, D.M. The Dawn of the Age of Amino Acid Sensors for the mTORC1 Pathway. Cell Metab. 2017, 26, 301–309.
- Zoncu, R.; Bar-Peled, L.; Efeyan, A.; Wang, S.; Sancak, Y.; Sabatini, D.M. mTORC1 Senses Lysosomal Amino Acids through an Inside-out Mechanism That Requires the Vacuolar H(+)-ATPase. Science 2011, 334, 678–683.
- Arnoult, D.; Grodet, A.; Lee, Y.-J.; Estaquier, J.; Blackstone, C. Release of OPA1 during Apoptosis Participates in the Rapid and Complete Release of Cytochrome c and Subsequent Mitochondrial Fragmentation. J. Biol. Chem. 2005, 280, 35742–35750.
- Arnoult, D.; Rismanchi, N.; Grodet, A.; Roberts, R.G.; Seeburg, D.P.; Estaquier, J.; Sheng, M.; Blackstone, C. Bax/Bak-Dependent Release of DDP/TIMM8a Promotes Drp1-Mediated Mitochondrial Fission and Mitoptosis during Programmed Cell Death. Curr. Biol. 2005, 15, 2112–2118.
- Tezze, C.; Romanello, V.; Desbats, M.A.; Fadini, G.P.; Albiero, M.; Favaro, G.; Ciciliot, S.; Soriano, M.E.; Morbidoni, V.; Cerqua, C.; et al. Age-Associated Loss of OPA1 in Muscle Impacts Muscle Mass, Metabolic Homeostasis, Systemic Inflammation, and Epithelial Senescence. Cell Metab. 2017, 25, 1374–1389.e6.
- Estaquier, J.; Arnoult, D. Inhibiting Drp1-Mediated Mitochondrial Fission Selectively Prevents the Release of Cytochrome c during Apoptosis. Cell Death Differ. 2007, 14, 1086–1094.
- Steffen, J.; Ngo, J.; Wang, S.-P.; Williams, K.; Kramer, H.F.; Ho, G.; Rodriguez, C.; Yekkala, K.; Amuzie, C.; Bialecki, R.; et al. The Mitochondrial Fission Protein Drp1 in Liver Is Required to Mitigate NASH and Prevents the Activation of the Mitochondrial ISR. Mol. Metab. 2022, 64, 101566.
- Ehses, S.; Raschke, I.; Mancuso, G.; Bernacchia, A.; Geimer, S.; Tondera, D.; Martinou, J.-C.; Westermann, B.; Rugarli, E.I.; Langer, T. Regulation of OPA1 Processing and Mitochondrial Fusion by M-AAA Protease Isoenzymes and OMA1. J. Cell Biol. 2009, 187, 1023–1036.
- Head, B.; Griparic, L.; Amiri, M.; Gandre-Babbe, S.; van der Bliek, A.M. Inducible Proteolytic Inactivation of OPA1 Mediated by the OMA1 Protease in Mammalian Cells. J. Cell Biol. 2009, 187, 959–966.
- Yang, J.; Baron, K.R.; Pride, D.E.; Schneemann, A.; Guo, X.; Chen, W.; Song, A.S.; Aviles, G.; Kampmann, M.; Luke Wiseman, R.; et al. DELE1 Oligomerization Promotes Integrated Stress Response Activation. Nat. Struct. Mol. Biol. 2023, 30, 1295–1302.
- Schubert, U.; Antón, L.C.; Bacík, I.; Cox, J.H.; Bour, S.; Bennink, J.R.; Orlowski, M.; Strebel, K.; Yewdell, J.W. CD4 Glycoprotein Degradation Induced by Human Immunodeficiency Virus Type 1 Vpu Protein Requires the Function of Proteasomes and the Ubiquitin-Conjugating Pathway. J. Virol. 1998, 72, 2280–2288.
- Butticaz, C.; Michielin, O.; Wyniger, J.; Telenti, A.; Rothenberger, S. Silencing of Both Beta-TrCP1 and HOS (Beta-TrCP2) Is Required to Suppress Human Immunodeficiency Virus Type 1 Vpu-Mediated CD4 down-Modulation. J. Virol. 2007, 81, 1502–1505.
- Douglas, J.L.; Viswanathan, K.; McCarroll, M.N.; Gustin, J.K.; Früh, K.; Moses, A.V. Vpu Directs the Degradation of the Human Immunodeficiency Virus Restriction Factor BST-2/Tetherin via a TrCP-Dependent Mechanism. J. Virol. 2009, 83, 7931–7947.
- Mangeat, B.; Gers-Huber, G.; Lehmann, M.; Zufferey, M.; Luban, J.; Piguet, V. HIV-1 Vpu Neutralizes the Antiviral Factor Tetherin/BST-2 by Binding It and Directing Its Beta-TrCP2-Dependent Degradation. PLoS Pathog. 2009, 5, e1000574.
- Margottin, F.; Bour, S.P.; Durand, H.; Selig, L.; Benichou, S.; Richard, V.; Thomas, D.; Strebel, K.; Benarous, R. A Novel Human WD Protein, h-Beta TrCp, That Interacts with HIV-1 Vpu Connects CD4 to the ER Degradation Pathway through an F-Box Motif. Mol. Cell 1998, 1, 565–574.
- Iwabu, Y.; Fujita, H.; Kinomoto, M.; Kaneko, K.; Ishizaka, Y.; Tanaka, Y.; Sata, T.; Tokunaga, K. HIV-1 Accessory Protein Vpu Internalizes Cell-Surface BST-2/Tetherin through Transmembrane Interactions Leading to Lysosomes. J. Biol. Chem. 2009, 284, 35060–35072.
- Mitchell, R.S.; Katsura, C.; Skasko, M.A.; Fitzpatrick, K.; Lau, D.; Ruiz, A.; Stephens, E.B.; Margottin-Goguet, F.; Benarous, R.; Guatelli, J.C. Vpu Antagonizes BST-2-Mediated Restriction of HIV-1 Release via Beta-TrCP and Endo-Lysosomal Trafficking. PLoS Pathog. 2009, 5, e1000450.
- Stoneham, C.A.; Singh, R.; Jia, X.; Xiong, Y.; Guatelli, J. Endocytic Activity of HIV-1 Vpu: Phosphoserine-Dependent Interactions with Clathrin Adaptors. Traffic 2017, 18, 545–561.
- Goffinet, C.; Allespach, I.; Homann, S.; Tervo, H.-M.; Habermann, A.; Rupp, D.; Oberbremer, L.; Kern, C.; Tibroni, N.; Welsch, S.; et al. HIV-1 Antagonism of CD317 Is Species Specific and Involves Vpu-Mediated Proteasomal Degradation of the Restriction Factor. Cell Host Microbe 2009, 5, 285–297.
- Lassot, I.; Ségéral, E.; Berlioz-Torrent, C.; Durand, H.; Groussin, L.; Hai, T.; Benarous, R.; Margottin-Goguet, F. ATF4 Degradation Relies on a Phosphorylation-Dependent Interaction with the SCF(betaTrCP) Ubiquitin Ligase. Mol. Cell Biol. 2001, 21, 2192–2202.
- Besnard-Guerin, C.; Belaïdouni, N.; Lassot, I.; Segeral, E.; Jobart, A.; Marchal, C.; Benarous, R. HIV-1 Vpu Sequesters Beta-Transducin Repeat-Containing Protein (betaTrCP) in the Cytoplasm and Provokes the Accumulation of Beta-Catenin and Other SCFbetaTrCP Substrates. J. Biol. Chem. 2004, 279, 788–795.
- Putters, J.; Slotman, J.A.; Gerlach, J.P.; Strous, G.J. Specificity, Location and Function of βTrCP Isoforms and Their Splice Variants. Cell Signal 2011, 23, 641–647.
- Pickering, S.; Sumner, J.; Kerridge, C.; Perera, M.; Neil, S. Differential Dysregulation of β-TrCP1 and -2 by HIV-1 Vpu Leads to Inhibition of Canonical and Non-Canonical NF-κB Pathways in Infected Cells. mBio 2023, 14, e0329322.
- Dalakas, M.C.; Illa, I.; Pezeshkpour, G.H.; Laukaitis, J.P.; Cohen, B.; Griffin, J.L. Mitochondrial Myopathy Caused by Long-Term Zidovudine Therapy. N. Engl. J. Med. 1990, 322, 1098–1105.
- Lewis, W.; Dalakas, M.C. Mitochondrial Toxicity of Antiviral Drugs. Nat. Med. 1995, 1, 417–422.
- Gerschenson, M.; Poirier, M.C. Fetal Patas Monkeys Sustain Mitochondrial Toxicity as a Result of in Utero Zidovudine Exposure. Ann. N. Y. Acad. Sci. 2000, 918, 269–281.
- Petit, F.; Fromenty, B.; Owen, A.; Estaquier, J. Mitochondria Are Sensors for HIV Drugs. Trends Pharmacol. Sci. 2005, 26, 258–264.
- Estaquier, J.; Lelièvre, J.-D.; Petit, F.; Brunner, T.; Moutouh-De Parseval, L.; Richman, D.D.; Ameisen, J.C.; Corbeil, J. Effects of Antiretroviral Drugs on Human Immunodeficiency Virus Type 1-Induced CD4(+) T-Cell Death. J. Virol. 2002, 76, 5966–5973.
- De Gassart, A.; Bujisic, B.; Zaffalon, L.; Decosterd, L.A.; Di Micco, A.; Frera, G.; Tallant, R.; Martinon, F. An Inhibitor of HIV-1 Protease Modulates Constitutive eIF2α Dephosphorylation to Trigger a Specific Integrated Stress Response. Proc. Natl. Acad. Sci. USA 2016, 113, E117–E126.
- Subeha, M.R.; Goyeneche, A.A.; Bustamante, P.; Lisio, M.A.; Burnier, J.V.; Telleria, C.M. Nelfinavir Induces Cytotoxicity towards High-Grade Serous Ovarian Cancer Cells, Involving Induction of the Unfolded Protein Response, Modulation of Protein Synthesis, DNA Damage, Lysosomal Impairment, and Potentiation of Toxicity Caused by Proteasome Inhibition. Cancers 2021, 14, 99.
- Brüning, A.; Rahmeh, M.; Friese, K. Nelfinavir and Bortezomib Inhibit mTOR Activity via ATF4-Mediated Sestrin-2 Regulation. Mol. Oncol. 2013, 7, 1012–1018.
- Liu, R.; Zhang, L.; Yang, J.; Zhang, X.; Mikkelsen, R.; Song, S.; Zhou, H. HIV Protease Inhibitors Sensitize Human Head and Neck Squamous Carcinoma Cells to Radiation by Activating Endoplasmic Reticulum Stress. PLoS ONE 2015, 10, e0125928.
- Fraichard, C.; Bonnet-Serrano, F.; Laguillier-Morizot, C.; Hebert-Schuster, M.; Lai-Kuen, R.; Sibiude, J.; Fournier, T.; Cohen, M.; Guibourdenche, J. Protease Inhibitor Anti-HIV, Lopinavir, Impairs Placental Endocrine Function. Int. J. Mol. Sci. 2021, 22, 683.
- McIntyre, R.L.; Molenaars, M.; Schomakers, B.V.; Gao, A.W.; Kamble, R.; Jongejan, A.; van Weeghel, M.; van Kuilenburg, A.B.P.; Possemato, R.; Houtkooper, R.H.; et al. Anti-Retroviral Treatment with Zidovudine Alters Pyrimidine Metabolism, Reduces Translation, and Extends Healthy Longevity via ATF-4. Cell Rep. 2023, 42, 111928.
- Barbieri, A.M.; Chiodini, I.; Ragni, E.; Colaianni, G.; Gadda, F.; Locatelli, M.; Lampertico, P.; Spada, A.; Eller-Vainicher, C. Suppressive Effects of Tenofovir Disoproxil Fumarate, an Antiretroviral Prodrug, on Mineralization and Type II and Type III Sodium-Dependent Phosphate Transporters Expression in Primary Human Osteoblasts. J. Cell Biochem. 2018, 119, 4855–4866.
- Ikebe, E.; Matsuoka, S.; Tezuka, K.; Kuramitsu, M.; Okuma, K.; Nakashima, M.; Kobayashi, S.; Makiyama, J.; Yamagishi, M.; Oyadomari, S.; et al. Activation of PERK-ATF4-CHOP Pathway as a Novel Therapeutic Approach for Efficient Elimination of HTLV-1-Infected Cells. Blood Adv. 2020, 4, 1845–1858.
- Lee, S.-D.; Yu, K.-L.; Park, S.-H.; Jung, Y.-M.; Kim, M.-J.; You, J.-C. Understanding of the Functional Role(s) of the Activating Transcription Factor 4(ATF4) in HIV Regulation and Production. BMB Rep. 2018, 51, 388–393.
- Vallejo-Gracia, A.; Chen, I.P.; Perrone, R.; Besnard, E.; Boehm, D.; Battivelli, E.; Tezil, T.; Krey, K.; Raymond, K.A.; Hull, P.A.; et al. FOXO1 Promotes HIV Latency by Suppressing ER Stress in T Cells. Nat. Microbiol. 2020, 5, 1144–1157.
- Xu, Y.; Weatherall, C.; Bailey, M.; Alcantara, S.; De Rose, R.; Estaquier, J.; Wilson, K.; Suzuki, K.; Corbeil, J.; Cooper, D.A.; et al. Simian Immunodeficiency Virus Infects Follicular Helper CD4 T Cells in Lymphoid Tissues during Pathogenic Infection of Pigtail Macaques. J. Virol. 2013, 87, 3760–3773.
- Moukambi, F.; Rabezanahary, H.; Rodrigues, V.; Racine, G.; Robitaille, L.; Krust, B.; Andreani, G.; Soundaramourty, C.; Silvestre, R.; Laforge, M.; et al. Early Loss of Splenic Tfh Cells in SIV-Infected Rhesus Macaques. PLoS Pathog. 2015, 11, e1005287.
- Rabezanahary, H.; Moukambi, F.; Palesch, D.; Clain, J.; Racine, G.; Andreani, G.; Benmadid-Laktout, G.; Zghidi-Abouzid, O.; Soundaramourty, C.; Tremblay, C.; et al. Despite Early Antiretroviral Therapy Effector Memory and Follicular Helper CD4 T Cells Are Major Reservoirs in Visceral Lymphoid Tissues of SIV-Infected Macaques. Mucosal Immunol. 2020, 13, 149–160.
- Banga, R.; Procopio, F.A.; Noto, A.; Pollakis, G.; Cavassini, M.; Ohmiti, K.; Corpataux, J.-M.; de Leval, L.; Pantaleo, G.; Perreau, M. PD-1(+) and Follicular Helper T Cells Are Responsible for Persistent HIV-1 Transcription in Treated Aviremic Individuals. Nat. Med. 2016, 22, 754–761.
- Perreau, M.; Savoye, A.-L.; De Crignis, E.; Corpataux, J.-M.; Cubas, R.; Haddad, E.K.; De Leval, L.; Graziosi, C.; Pantaleo, G. Follicular Helper T Cells Serve as the Major CD4 T Cell Compartment for HIV-1 Infection, Replication, and Production. J. Exp. Med. 2013, 210, 143–156.
- Pluta, A.; Jaworski, J.P.; Cortés-Rubio, C.N. Balance between Retroviral Latency and Transcription: Based on HIV Model. Pathogens 2020, 10, 16.
- Reddy, T.R.; Tang, H.; Li, X.; Wong-Staal, F. Functional Interaction of the HTLV-1 Transactivator Tax with Activating Transcription Factor-4 (ATF4). Oncogene 1997, 14, 2785–2792.
- Pereira, L.A.; Bentley, K.; Peeters, A.; Churchill, M.J.; Deacon, N.J. A Compilation of Cellular Transcription Factor Interactions with the HIV-1 LTR Promoter. Nucleic Acids Res. 2000, 28, 663–668.
- Rabbi, M.F.; Saifuddin, M.; Gu, D.S.; Kagnoff, M.F.; Roebuck, K.A. U5 Region of the Human Immunodeficiency Virus Type 1 Long Terminal Repeat Contains TRE-like cAMP-Responsive Elements That Bind Both AP-1 and CREB/ATF Proteins. Virology 1997, 233, 235–245.
- Neill, G.; Masson, G.R. A Stay of Execution: ATF4 Regulation and Potential Outcomes for the Integrated Stress Response. Front. Mol. Neurosci. 2023, 16, 1112253.
- Rodríguez-Martínez, J.A.; Reinke, A.W.; Bhimsaria, D.; Keating, A.E.; Ansari, A.Z. Combinatorial bZIP Dimers Display Complex DNA-Binding Specificity Landscapes. eLife 2017, 6, e19272.
- Hai, T.; Curran, T. Cross-Family Dimerization of Transcription Factors Fos/Jun and ATF/CREB Alters DNA Binding Specificity. Proc. Natl. Acad. Sci. USA 1991, 88, 3720–3724.
- Mu, Y.; Yu, Y.; Yue, X.; Musarat, I.; Gong, R.; Zhu, C.; Liu, Y.; Liu, F.; Zhu, Y.; Wu, J. The X Protein of HBV Induces HIV-1 Long Terminal Repeat Transcription by Enhancing the Binding of C/EBPβ and CREB1/2 Regulatory Proteins to the Long Terminal Repeat of HIV-1. Virus Res. 2011, 156, 81–90.
- Gachon, F.; Thebault, S.; Peleraux, A.; Devaux, C.; Mesnard, J.M. Molecular Interactions Involved in the Transactivation of the Human T-Cell Leukemia Virus Type 1 Promoter Mediated by Tax and CREB-2 (ATF-4). Mol. Cell Biol. 2000, 20, 3470–3481.
- Ameisen, J.C.; Estaquier, J.; Idziorek, T. From AIDS to Parasite Infection: Pathogen-Mediated Subversion of Programmed Cell Death as a Mechanism for Immune Dysregulation. Immunol. Rev. 1994, 142, 9–51.
- Clerici, M.; Sarin, A.; Coffman, R.L.; Wynn, T.A.; Blatt, S.P.; Hendrix, C.W.; Wolf, S.F.; Shearer, G.M.; Henkart, P.A. Type 1/Type 2 Cytokine Modulation of T-Cell Programmed Cell Death as a Model for Human Immunodeficiency Virus Pathogenesis. Proc. Natl. Acad. Sci. USA 1994, 91, 11811–11815.
- Finkel, T.H.; Tudor-Williams, G.; Banda, N.K.; Cotton, M.F.; Curiel, T.; Monks, C.; Baba, T.W.; Ruprecht, R.M.; Kupfer, A. Apoptosis Occurs Predominantly in Bystander Cells and Not in Productively Infected Cells of HIV- and SIV-Infected Lymph Nodes. Nat. Med. 1995, 1, 129–134.
- Arnoult, D.; Viollet, L.; Petit, F.; Lelièvre, J.-D.; Estaquier, J. HIV-1 Triggers Mitochondrion Death. Mitochondrion 2004, 4, 255–269.
- Estaquier, J.; Tanaka, M.; Suda, T.; Nagata, S.; Golstein, P.; Ameisen, J.C. Fas-Mediated Apoptosis of CD4+ and CD8+ T Cells from Human Immunodeficiency Virus-Infected Persons: Differential in Vitro Preventive Effect of Cytokines and Protease Antagonists. Blood 1996, 87, 4959–4966.
- Estaquier, J.; Idziorek, T.; Zou, W.; Emilie, D.; Farber, C.M.; Bourez, J.M.; Ameisen, J.C. T Helper Type 1/T Helper Type 2 Cytokines and T Cell Death: Preventive Effect of Interleukin 12 on Activation-Induced and CD95 (FAS/APO-1)-Mediated Apoptosis of CD4+ T Cells from Human Immunodeficiency Virus-Infected Persons. J. Exp. Med. 1995, 182, 1759–1767.
- Katsikis, P.D.; Wunderlich, E.S.; Smith, C.A.; Herzenberg, L.A.; Herzenberg, L.A. Fas Antigen Stimulation Induces Marked Apoptosis of T Lymphocytes in Human Immunodeficiency Virus-Infected Individuals. J. Exp. Med. 1995, 181, 2029–2036.
- Sloand, E.M.; Young, N.S.; Kumar, P.; Weichold, F.F.; Sato, T.; Maciejewski, J.P. Role of Fas Ligand and Receptor in the Mechanism of T-Cell Depletion in Acquired Immunodeficiency Syndrome: Effect on CD4+ Lymphocyte Depletion and Human Immunodeficiency Virus Replication. Blood 1997, 89, 1357–1363.
- Galehdar, Z.; Swan, P.; Fuerth, B.; Callaghan, S.M.; Park, D.S.; Cregan, S.P. Neuronal Apoptosis Induced by Endoplasmic Reticulum Stress Is Regulated by ATF4-CHOP-Mediated Induction of the Bcl-2 Homology 3-Only Member PUMA. J. Neurosci. 2010, 30, 16938–16948.
- Puthalakath, H.; O’Reilly, L.A.; Gunn, P.; Lee, L.; Kelly, P.N.; Huntington, N.D.; Hughes, P.D.; Michalak, E.M.; McKimm-Breschkin, J.; Motoyama, N.; et al. ER Stress Triggers Apoptosis by Activating BH3-Only Protein Bim. Cell 2007, 129, 1337–1349.
- Bagheri-Yarmand, R.; Sinha, K.M.; Gururaj, A.E.; Ahmed, Z.; Rizvi, Y.Q.; Huang, S.-C.; Ladbury, J.E.; Bogler, O.; Williams, M.D.; Cote, G.J.; et al. A Novel Dual Kinase Function of the RET Proto-Oncogene Negatively Regulates Activating Transcription Factor 4-Mediated Apoptosis. J. Biol. Chem. 2015, 290, 11749–11761.
- Teske, B.F.; Fusakio, M.E.; Zhou, D.; Shan, J.; McClintick, J.N.; Kilberg, M.S.; Wek, R.C. CHOP Induces Activating Transcription Factor 5 (ATF5) to Trigger Apoptosis in Response to Perturbations in Protein Homeostasis. Mol. Biol. Cell 2013, 24, 2477–2490.
- Wang, Q.; Mora-Jensen, H.; Weniger, M.A.; Perez-Galan, P.; Wolford, C.; Hai, T.; Ron, D.; Chen, W.; Trenkle, W.; Wiestner, A.; et al. ERAD Inhibitors Integrate ER Stress with an Epigenetic Mechanism to Activate BH3-Only Protein NOXA in Cancer Cells. Proc. Natl. Acad. Sci. USA 2009, 106, 2200–2205.
- Hiramatsu, N.; Messah, C.; Han, J.; LaVail, M.M.; Kaufman, R.J.; Lin, J.H. Translational and Posttranslational Regulation of XIAP by eIF2α and ATF4 Promotes ER Stress-Induced Cell Death during the Unfolded Protein Response. Mol. Biol. Cell 2014, 25, 1411–1420.
- B’chir, W.; Maurin, A.-C.; Carraro, V.; Averous, J.; Jousse, C.; Muranishi, Y.; Parry, L.; Stepien, G.; Fafournoux, P.; Bruhat, A. The eIF2α/ATF4 Pathway Is Essential for Stress-Induced Autophagy Gene Expression. Nucleic Acids Res. 2013, 41, 7683–7699.
- Whitney, M.L.; Jefferson, L.S.; Kimball, S.R. ATF4 Is Necessary and Sufficient for ER Stress-Induced Upregulation of REDD1 Expression. Biochem. Biophys. Res. Commun. 2009, 379, 451–455.
- Wolff, N.C.; Vega-Rubin-de-Celis, S.; Xie, X.-J.; Castrillon, D.H.; Kabbani, W.; Brugarolas, J. Cell-Type-Dependent Regulation of mTORC1 by REDD1 and the Tumor Suppressors TSC1/TSC2 and LKB1 in Response to Hypoxia. Mol. Cell Biol. 2011, 31, 1870–1884.
- Xu, D.; Dai, W.; Kutzler, L.; Lacko, H.A.; Jefferson, L.S.; Dennis, M.D.; Kimball, S.R. ATF4-Mediated Upregulation of REDD1 and Sestrin2 Suppresses mTORC1 Activity during Prolonged Leucine Deprivation. J. Nutr. 2020, 150, 1022–1030.
- Cabrera-Rodríguez, R.; Pérez-Yanes, S.; Lorenzo-Sánchez, I.; Trujillo-González, R.; Estévez-Herrera, J.; García-Luis, J.; Valenzuela-Fernández, A. HIV Infection: Shaping the Complex, Dynamic, and Interconnected Network of the Cytoskeleton. Int. J. Mol. Sci. 2023, 24, 13104.
- Dinkins, C.; Pilli, M.; Kehrl, J.H. Roles of Autophagy in HIV Infection. Immunol. Cell Biol. 2015, 93, 11–17.
- Espert, L.; Denizot, M.; Grimaldi, M.; Robert-Hebmann, V.; Gay, B.; Varbanov, M.; Codogno, P.; Biard-Piechaczyk, M. Autophagy Is Involved in T Cell Death after Binding of HIV-1 Envelope Proteins to CXCR4. J. Clin. Investig. 2006, 116, 2161–2172.
- García-Expósito, L.; Barroso-González, J.; Puigdomènech, I.; Machado, J.-D.; Blanco, J.; Valenzuela-Fernández, A. HIV-1 Requires Arf6-Mediated Membrane Dynamics to Efficiently Enter and Infect T Lymphocytes. Mol. Biol. Cell 2011, 22, 1148–1166.
- Moreau, K.; Ravikumar, B.; Puri, C.; Rubinsztein, D.C. Arf6 Promotes Autophagosome Formation via Effects on Phosphatidylinositol 4,5-Bisphosphate and Phospholipase D. J. Cell Biol. 2012, 196, 483–496.
- Van Grol, J.; Subauste, C.; Andrade, R.M.; Fujinaga, K.; Nelson, J.; Subauste, C.S. HIV-1 Inhibits Autophagy in Bystander Macrophage/Monocytic Cells through Src-Akt and STAT3. PLoS ONE 2010, 5, e11733.
- Zhou, D.; Spector, S.A. Human Immunodeficiency Virus Type-1 Infection Inhibits Autophagy. AIDS 2008, 22, 695–699.
- Alfaisal, J.; Machado, A.; Galais, M.; Robert-Hebmann, V.; Arnauné-Pelloquin, L.; Espert, L.; Biard-Piechaczyk, M. HIV-1 Vpr Inhibits Autophagy during the Early Steps of Infection of CD4 T Cells. Biol. Cell 2019, 111, 308–318.
- Laforge, M.; Limou, S.; Harper, F.; Casartelli, N.; Rodrigues, V.; Silvestre, R.; Haloui, H.; Zagury, J.-F.; Senik, A.; Estaquier, J. DRAM Triggers Lysosomal Membrane Permeabilization and Cell Death in CD4(+) T Cells Infected with HIV. PLoS Pathog. 2013, 9, e1003328.
- Rozman, M.; Zidovec-Lepej, S.; Jambrosic, K.; Babić, M.; Drmić Hofman, I. Role of TLRs in HIV-1 Infection and Potential of TLR Agonists in HIV-1 Vaccine Development and Treatment Strategies. Pathogens 2023, 12, 92.
- Ning, S.; Pagano, J.S.; Barber, G.N. IRF7: Activation, Regulation, Modification and Function. Genes Immun. 2011, 12, 399–414.
- Liang, Q.; Deng, H.; Sun, C.-W.; Townes, T.M.; Zhu, F. Negative Regulation of IRF7 Activation by Activating Transcription Factor 4 Suggests a Cross-Regulation between the IFN Responses and the Cellular Integrated Stress Responses. J. Immunol. 2011, 186, 1001–1010.
- Farook, J.M.; Shields, J.; Tawfik, A.; Markand, S.; Sen, T.; Smith, S.B.; Brann, D.; Dhandapani, K.M.; Sen, N. GADD34 Induces Cell Death through Inactivation of Akt Following Traumatic Brain Injury. Cell Death Dis. 2013, 4, e754.
- Imbeault, M.; Ouellet, M.; Tremblay, M.J. Microarray Study Reveals That HIV-1 Induces Rapid Type-I Interferon-Dependent P53 mRNA up-Regulation in Human Primary CD4+ T Cells. Retrovirology 2009, 6, 5.
- Sirois, M.; Robitaille, L.; Allary, R.; Shah, M.; Woelk, C.H.; Estaquier, J.; Corbeil, J. TRAF6 and IRF7 Control HIV Replication in Macrophages. PLoS ONE 2011, 6, e28125.
- Ishaq, M.; Marshall, H.; Natarajan, V. GADD34 Attenuates HIV-1 Replication by Viral 5′-UTR TAR RNA-Mediated Translational Inhibition. Virology 2020, 540, 119–131.
- Guo, H.; Wang, Q.; Ghneim, K.; Wang, L.; Rampanelli, E.; Holley-Guthrie, E.; Cheng, L.; Garrido, C.; Margolis, D.M.; Eller, L.A.; et al. Multi-Omics Analyses Reveal That HIV-1 Alters CD4+ T Cell Immunometabolism to Fuel Virus Replication. Nat. Immunol. 2021, 22, 423–433.
- Palmer, C.S.; Ostrowski, M.; Gouillou, M.; Tsai, L.; Yu, D.; Zhou, J.; Henstridge, D.C.; Maisa, A.; Hearps, A.C.; Lewin, S.R.; et al. Increased Glucose Metabolic Activity Is Associated with CD4+ T-Cell Activation and Depletion during Chronic HIV Infection. AIDS 2014, 28, 297–309.
- Valle-Casuso, J.C.; Angin, M.; Volant, S.; Passaes, C.; Monceaux, V.; Mikhailova, A.; Bourdic, K.; Avettand-Fenoel, V.; Boufassa, F.; Sitbon, M.; et al. Cellular Metabolism Is a Major Determinant of HIV-1 Reservoir Seeding in CD4+ T Cells and Offers an Opportunity to Tackle Infection. Cell Metab. 2019, 29, 611–626.e5.
- Loisel-Meyer, S.; Swainson, L.; Craveiro, M.; Oburoglu, L.; Mongellaz, C.; Costa, C.; Martinez, M.; Cosset, F.-L.; Battini, J.-L.; Herzenberg, L.A.; et al. Glut1-Mediated Glucose Transport Regulates HIV Infection. Proc. Natl. Acad. Sci. USA 2012, 109, 2549–2554.
- Yang, X.; Xia, R.; Yue, C.; Zhai, W.; Du, W.; Yang, Q.; Cao, H.; Chen, X.; Obando, D.; Zhu, Y.; et al. ATF4 Regulates CD4+ T Cell Immune Responses through Metabolic Reprogramming. Cell Rep. 2018, 23, 1754–1766.
- Clerc, I.; Moussa, D.A.; Vahlas, Z.; Tardito, S.; Oburoglu, L.; Hope, T.J.; Sitbon, M.; Dardalhon, V.; Mongellaz, C.; Taylor, N. Entry of Glucose- and Glutamine-Derived Carbons into the Citric Acid Cycle Supports Early Steps of HIV-1 Infection in CD4 T Cells. Nat. Metab. 2019, 1, 717–730.
- Yero, A.; Bouassa, R.-S.M.; Ancuta, P.; Estaquier, J.; Jenabian, M.-A. Immuno-Metabolic Control of the Balance between Th17-Polarized and Regulatory T-Cells during HIV Infection. Cytokine Growth Factor Rev. 2023, 69, 1–13.
- Sundrud, M.S.; Koralov, S.B.; Feuerer, M.; Calado, D.P.; Kozhaya, A.E.; Rhule-Smith, A.; Lefebvre, R.E.; Unutmaz, D.; Mazitschek, R.; Waldner, H.; et al. Halofuginone Inhibits TH17 Cell Differentiation by Activating the Amino Acid Starvation Response. Science 2009, 324, 1334–1338.
- Gosselin, A.; Monteiro, P.; Chomont, N.; Diaz-Griffero, F.; Said, E.A.; Fonseca, S.; Wacleche, V.; El-Far, M.; Boulassel, M.-R.; Routy, J.-P.; et al. Peripheral Blood CCR4+CCR6+ and CXCR3+CCR6+CD4+ T Cells Are Highly Permissive to HIV-1 Infection. J. Immunol. 2010, 184, 1604–1616.
- Gosselin, A.; Wiche Salinas, T.R.; Planas, D.; Wacleche, V.S.; Zhang, Y.; Fromentin, R.; Chomont, N.; Cohen, É.A.; Shacklett, B.; Mehraj, V.; et al. HIV Persists in CCR6+CD4+ T Cells from Colon and Blood during Antiretroviral Therapy. AIDS 2017, 31, 35–48.
- Estes, J.D.; Harris, L.D.; Klatt, N.R.; Tabb, B.; Pittaluga, S.; Paiardini, M.; Barclay, G.R.; Smedley, J.; Pung, R.; Oliveira, K.M.; et al. Damaged Intestinal Epithelial Integrity Linked to Microbial Translocation in Pathogenic Simian Immunodeficiency Virus Infections. PLoS Pathog. 2010, 6, e1001052.
- Favre, D.; Lederer, S.; Kanwar, B.; Ma, Z.-M.; Proll, S.; Kasakow, Z.; Mold, J.; Swainson, L.; Barbour, J.D.; Baskin, C.R.; et al. Critical Loss of the Balance between Th17 and T Regulatory Cell Populations in Pathogenic SIV Infection. PLoS Pathog. 2009, 5, e1000295.
- McGary, C.S.; Alvarez, X.; Harrington, S.; Cervasi, B.; Ryan, E.S.; Iriele, R.I.; Paganini, S.; Harper, J.L.; Easley, K.; Silvestri, G.; et al. The Loss of CCR6+ and CD161+ CD4+ T-Cell Homeostasis Contributes to Disease Progression in SIV-Infected Rhesus Macaques. Mucosal Immunol. 2017, 10, 1082–1096.
- Raffatellu, M.; Santos, R.L.; Verhoeven, D.E.; George, M.D.; Wilson, R.P.; Winter, S.E.; Godinez, I.; Sankaran, S.; Paixao, T.A.; Gordon, M.A.; et al. Simian Immunodeficiency Virus-Induced Mucosal Interleukin-17 Deficiency Promotes Salmonella Dissemination from the Gut. Nat. Med. 2008, 14, 421–428.
- Yero, A.; Farnos, O.; Rabezanahary, H.; Racine, G.; Estaquier, J.; Jenabian, M.-A. Differential Dynamics of Regulatory T-Cell and Th17 Cell Balance in Mesenteric Lymph Nodes and Blood Following Early Antiretroviral Initiation during Acute Simian Immunodeficiency Virus Infection. J. Virol. 2019, 93, e00371-19.
More
Information
Subjects:
Cell Biology; Virology; Immunology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
688
Revisions:
2 times
(View History)
Update Date:
18 Mar 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




