Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Wang, M.; Fan, X.; Ding, F. Jasmonate. Encyclopedia. Available online: https://encyclopedia.pub/entry/55797 (accessed on 11 January 2026).
Wang M, Fan X, Ding F. Jasmonate. Encyclopedia. Available at: https://encyclopedia.pub/entry/55797. Accessed January 11, 2026.
Wang, Meiling, Xiulan Fan, Fei Ding. "Jasmonate" Encyclopedia, https://encyclopedia.pub/entry/55797 (accessed January 11, 2026).
Wang, M., Fan, X., & Ding, F. (2024, March 04). Jasmonate. In Encyclopedia. https://encyclopedia.pub/entry/55797
Wang, Meiling, et al. "Jasmonate." Encyclopedia. Web. 04 March, 2024.
Copy Citation
Temperature is a critical environmental factor that plays a vital role in plant growth and development. Temperatures below or above the optimum ranges lead to cold or heat stress, respectively. Temperature stress retards plant growth and development, and it reduces crop yields. Jasmonates (JAs) are a class of oxylipin phytohormones that play various roles in growth, development, and stress response.
heat stress
temperature stress
jasmonates
1. Introduction
Plants are unable to move and thus have to cope with various adverse environmental factors, such as extreme temperatures, drought, salinity, and heavy metal toxicity [1][2]. Being a critical environmental factor, temperature plays a dominant role in plant growth and development [3], and it determines the geographical distribution of plant species [4]. As global climate change intensifies, the magnitude and frequency of extreme temperatures are increasing. Extreme temperatures cause various forms of damage at different stages during plant growth and development. The general consequences of heat and cold stress include impaired photosynthesis, excessive accumulation of reactive oxygen species, broken plasma membranes, and altered phytohormone levels [5][6][7][8][9][10][11]. Eventually, heat and cold stress inhibit plant growth and cause losses in crop yields, posing a serious threat to food security [12][13][14][15]. A survey showed that wheat, rice, maize, and soybean yields would decrease by 6.0%, 3.2%, 7.4%, and 3.1% on average, respectively, if the global temperature rises by 1 °C [4][16]. Low temperatures adversely affect plant growth and development and reduce crop production [12][17][18][19]. For instance, it has been estimated that in temperate and high-elevation regions, cold stress accounts for 30–40% of yield losses in rice [4][20].
Unlike animals, plants are sessile organisms and thus are unable to escape unfavorable temperature conditions. Instead, plants have evolved a set of sophisticated strategies enabling them to survive under temperature stress. Plant hormones play a vital role in the initiation of temperature stress response by integrating temperature stimulus and endogenous signals. For instance, jasmonates (JAs), abscisic acid (ABA), and brassinosteroid (BR) positively regulate plant response to both heat and cold stress [21][22][23][24][25][26]. Jasmonates (JAs) are a typical class of phytohormones. The term “jasmonates” generally refers to jasmonic acid and its derivatives, typically including jasmonyl isoleucine (JA-Ile) and methyl jasmonate (MeJA) [27]. In addition to its well-known role in growth and development, and in defense against pathogen attack and insect herbivory [28][29][30][31][32], a growing number of studies have highlighted the vital role of JA in the response to a variety of abiotic stresses, including drought, salinity, heat, and cold stress response [33][34][35][36][37][38][39][40][41][42][43].
2. JA Biosynthesis and Signaling
The JA biosynthetic pathway and the major enzymes involved have been well characterized and extensively reviewed [27][29][44][45][46][47][48]. JA biosynthesis starts with polyunsaturated fatty acids released from plastid membranes through the action of phospholipase (PLA) [49]. Current evidence supports the assertion that JA is derived via the α-linolenic acid (α-LeA, C18:3) pathway and the hexadecatrienoic acid (HTA, C16:3) pathway [50]. As the α-LeA pathway is dominant in the biosynthesis of JA, researchers focus on this pathway to explain JA biosynthesis (Figure 1A). Overall, four major enzymes are engaged in JA biosynthesis from α-LeA, comprising lipoxygenase (LOX), allene oxide synthase (AOS), allene oxide cyclase (AOC), and oxophytodienoic acid reductase (OPR) [51][52]. In plastids, LOX catalyzes the first step of JA biosynthesis. α-LeA is converted to 13(S)-hydroperoxy-octadecatrienoic acid (13-HPOT) by LOXs. In Arabidopsis, four LOXs—LOX2, LOX3, LOX4, and LOX6—are able to oxygenate α-LeA [53]. Each LOX may function differentially depending on the types of external stimuli. For instance, LOX6 is predominantly involved in JA production upon wounding and drought stress [54][55]. Subsequently, 13-HPOT is catalyzed by AOS to produce 12,13(S)-epoxy-octadecatrienoic acid (12,13-EOT). AOS is a cytochrome P450 enzyme, which uses oxygenated fatty acid hydroperoxide substrates as oxygen donors. There is only one AOS gene in Arabidopsis, and mutation in AOS leads to disrupted JA biosynthesis in response to wounding [56]. 12,13-EOT is further converted to 12-oxo-phytodienoic acid (12-OPDA), catalyzed by AOC. Four AOC genes have been identified and found to act redundantly in the biosynthesis of JA in Arabidopsis [57]. Next, 12-OPDA is translocated by the transporter COMATOSE (CTS1) to the peroxisome [58]. In the peroxisome, OPDA is reduced by OPDA reductase (OPR) to produce OPC-8:0. OPRs are encoded by six OPR genes in the Arabidopsis genome; however, only OPR3 acts on OPDA. OPC-8:0 is then subjected to three rounds of β-oxidation by acyl-CoA oxidase (ACX), L-3-KETOACYLCOA THIOLASE (KAT), and multifunctional protein (MFP) [59]. Finally, JasmonoylCoA, which is generated through β-oxidation reaction, can be further cleaved by THIOESTERASE (TE) to produce (+)-7-iso-JA, which is then transported to the cytoplasm. In the cytoplasm, various JA derivatives are formed, including methyl jasmonate (MeJA) and JA-isoleucine (JA-Ile) [60][61]. The conjugation of (+)-7-iso-JA with isoleucine produces JA-Ile, and the reaction is catalyzed by jasmonate-amido synthetase (JAR1). The jar1 mutant was identified as the first JA-insensitive mutant, and the JA-Ile level in mutant plants is severely reduced [62]. The methylation of JA forms MeJA, with catalysis by jasmonate methyl transferase (JMT). JA-Ile and MeJA are active forms of JA, and they can be converted to inactive 12-OH-JA by jasmonate-induced oxygenases (JO) and jasmonic acid oxidases (JAO) [63][64].
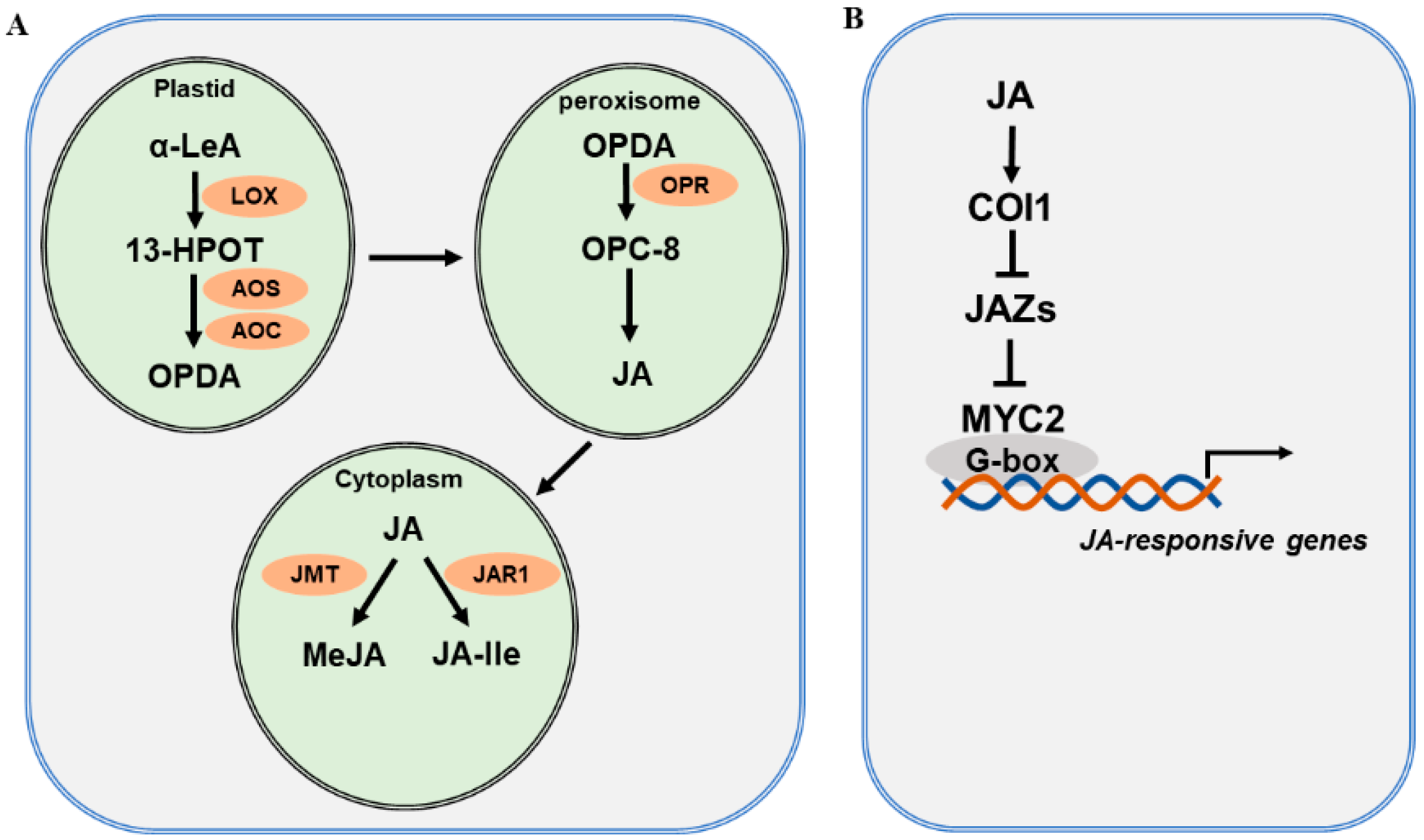
Figure 1. JA biosynthesis and signaling pathway. (A) A simplified JA biosynthesis pathway from α-linolenic acid (α-LeA). JA and its derivatives are produced from α-LeA through several sequential steps, which are catalyzed by lipoxygenase (LOX), allene oxide synthase (AOS), and allene oxide cyclase (AOC) in plastids; OPDA reductase (OPR) in peroxisomes; and jasmonate-amido synthetase (JAR1) and jasmonate methyl transferase (JMT) in the cytoplasm. (B) A simplified JA signaling pathway. Three main components are involved in JA signaling: the COI receptor, the JAZ repressor, and the MYC2 transcription factor.
The JA signaling pathway has been well defined primarily in Arabidopsis and tomato. In brief, it consists of the receptor CORONATINE INSENSITIVE 1 (COI1), the repressors JASMONATE ZIM-DOMAIN PROTEIN (JAZs), and the master transcription factors MYCs (Figure 1B). Early genetic screens identified coronatine insensitive 1 (coi1), which is insensitive to the functional homolog of JA-Ile, coronatine. Later studies confirmed that COI1 acts as a receptor that perceives active JA [65][66]. COI1 is an F-box protein [67] that is able to associate with SKP1 and CULLIN1 to form an E3 ubiquitin ligase complex, SCFCOI1. In search of the substrate for SCFCOI1, researchers from three independent laboratories discovered JASMONATE ZIM-DOMAIN (JAZ) proteins, which are repressors of JA signaling [68][69][70]. JAZs belong to the plant-specific TIFY family, possessing a core TIF[F/Y]XG motif within the ZIM (ZN-FINGER PROTEIN EXPRESSED IN INFLORESCENCE MERISTEM) protein domain. There are 12 JAZ proteins that have been identified in Arabidopsis [68][69][71]. These JAZs are distinct from other proteins in the TIFY family as they contain a C-terminal Jas motif, SLX2FX2KRX2RX5PY [70][72][73]. The interaction of COI1 with the Jas domain of JAZ proteins forms the co-receptor complex [74][75]. TOPLESS (TPL) and TPL-related proteins (TPRs) are corepressors that interact with JAZ proteins through the ETHYLENE RESPONSE FACTOR (ERF)-ASSOCIATED AMPHIPHILIC REPRESSION (EAR) motif. MYC2, a bHLH transcription factor, is the master regulator of JA signaling and mediates a variety of biological processes. MYC2 is repressed by JAZ proteins and is released following the degradation of JAZs [69][75][76]. Eventually, MYC2 activates various downstream JA-responsive genes [44][77][78][79]. MYC2 plays multifaceted roles in growth and development, defense against biotic stress, abiotic stress response, and regulation of secondary metabolite biosynthesis. MYC3 and MYC4 are two close homologs of MYC2. MYC2 forms dimers with MYC3 and MYC4 to modulate the transcription of various target genes by binding to the G-box or its variants within the promoters [80][81].
3. The Role of JA in Cold Stress Response
Cold stress generally refers to two types of stresses: chilling stress, with a temperature ranging from 0 °C to 15 °C, and freezing stress, with a temperature below 0 °C. Cold stress is one of the most severe environmental stresses in plants. Cold stress inhibits plant growth and development and threatens crop productivity. To cope with cold stress, plants have evolved a wide variety of mechanisms. JA, a classical phytohormone, positively mediates plant cold response. Plenty of studies have shown that JA production is increased in plants in response to cold stress, which implies the potential role of JA in the response to cold stress. For instance, upon cold stress, JA accumulation is markedly enhanced in Arabidopsis, tomato, and rice [82][83][84][85]. Consistent with increased JA accumulation, the expression of JA biosynthesis genes is induced by low temperatures. As observed in rice, cold stress triggers the expression of OsLOX2, OsAOC, OsAOS1, and OsAOS2 and promotes endogenous JA levels [85]. Similarly, in Artemisia annua, higher levels of JA and increased expression of JA biosynthesis genes were observed following cold treatment [35]. Furthermore, the application of exogenous MeJA potentiates cold tolerance in a variety of species, including banana, tomato, loquat, orange, guava, mango, and peach [24][86][87][88][89][90]. All these results support the assertion that JA is involved in plant cold stress response.
The role of JA in cold response is further substantiated by mutant or transgenic plants with altered JA biosynthesis. Arabidopsis plants with mutations in AOS and LOX2 show impaired JA biosynthesis, and these plants are hypersensitive to low temperatures [82]. Another study showed that MaLBD5 (lateral-organ boundaries domain) is associated with the JA-mediated cold response in banana fruits. MaLBD5 promotes JA biosynthesis by transactivating the expression of MaAOC2 [91]. Furthermore, a genetic study showed that HAN1, a rice gene that encodes an oxidase that catalyzes the active form JA-Ile to the inactive form 12OH-JAIle, negatively regulates cold tolerance [92]. In addition, transgenic Arabidopsis plants overexpressing GLR1.2 (glutamate-like receptor) and GLR1.3 display enhanced accumulation of JA by activating the expression of JA biosynthesis genes and increased cold tolerance [93].
In an attempt to understand the underlying mechanisms of JA-mediated cold tolerance, numerous studies have revealed that major components of the JA signaling pathway play a critical role in cold tolerance in plants. Being the master regulator of JA signaling, MYC2 is of great importance in cold response. In Poncirus trifoliata, MYC2 targets a betaine aldehyde dehydrogenase gene (PtrBADH-l) and directly upregulates it, thereby increasing the production of glycine betaine. A high level of glycine betaine confers cold tolerance in Poncirus trifoliata [94]. In tomato, MYC2 targets and upregulates ADC1, which is a putrescine biosynthesis gene, leading to enhanced putrescine accumulation and decreased cold damage [83]. Under cold conditions, MYC2 directly stimulates the expression of SlGSTU24, a JA-responsive glutathione S-transferase gene, and consequently alleviates cold-induced oxidative stress [84]. These results indicate that JA positively regulates cold response by promoting the production of antioxidant enzymes and non-enzymatic cryoprotective compounds through MYC2.
The module ICE (inducer of CBF expression)-DREB1/CBF (dehydration-response element-binding protein 1/C-repeat binding factors) plays a vital role in cold response in plants [95][96]. DREB1/CBFs are AP2/ERF (APETALA2/ETHYLENE-RESPONSIVE FACTOR)-type transcription factors capable of binding to DREs (dehydration-responsive element) in the promoters of target genes and acting as key regulators of COR (cold-regulated) genes [97][98][99]. Previous studies have identified three DREB1/CBF genes: DREB1A/CBF3, DREB1B/CBF1, and DREB1C/CBF2 [98][100]. Cold stress leads to rapid induction of these genes, and mutations in them severely impair cold tolerance [101][102]. Overexpression of CBF genes induces the expression of numerous cold-inducible genes and confers cold tolerance [103][104]. ICE1 is an MYC-like basic helix–loop–helix transcription factor that acts as a master regulator in the DREB1/CBF pathway. In the past two decades, a large number of studies have established the role of ICE1 in the expression of DREB1/CBF. However, recently, it has been reported that repression of CBF3 in ice1-1 mutant plants is due to DNA-methylation-mediated gene silencing caused by inserted T-DNA, not by ICE1 mutation, and that ICE1 is not associated with CBF3 expression [105][106][107].
JAZs, the repressors of JA signaling, are important for the JA-mediated cold response, and the ICE-CBF module is involved in this process. In Arabidopsis, JAZs interact with ICEs to repress the expression of CBFs. Upon cold treatment, JA accumulation is increased, promoting the degradation of JAZs, thus releasing ICEs. ICEs then activate CBFs, conferring cold tolerance in Arabidopsis [82]. In apple, MdJAZ1 and MdJAZ2 interact with the transcription factor BBX37 and suppress the transcription of MdCBF1 and MdCBF4. In response to cold stress, increased JA leads to the degradation of MdJAZ1 and MdJAZ2, allowing BBX37 to activate MdCBF1 and MdCBF4 [108]. Interestingly, under cold stress, the expression of MaMYC2a and MaMYC2b is tremendously induced in banana, and MaMYC2 physically interacts with MaICE1, thus triggering the expression of MaCBF1 and MaCBF2 [87]. In Arabidopsis, SFR6 (SENSITIVE TO FREEZING 6) controls the expression of cold-regulated genes by acting on the CBF module [109][110][111]. Meanwhile, SFR6 is also involved in the regulation of JA responses [112][113]. These studies highlight the role of JA in cold response via the induction of CBFs.
References
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324.
- Zhou, H.; Shi, H.; Yang, Y.; Feng, X.; Chen, X.; Xiao, F.; Lin, H.; Guo, Y. Insights into Plant Salt Stress Signaling and Tolerance. J. Genet. Genom. 2023, in press.
- Gao, Z.; Zhou, Y.; He, Y. Molecular Epigenetic Mechanisms for the Memory of Temperature Stresses in Plants. J. Genet. Genom. 2022, 49, 991–1001.
- Ding, Y.; Yang, S. Surviving and Thriving: How Plants Perceive and Respond to Temperature Stress. Dev. Cell 2022, 57, 947–958.
- Huang, J.; Zhao, X.; Bürger, M.; Chory, J.; Wang, X. The Role of Ethylene in Plant Temperature Stress Response. Trends Plant Sci. 2023, 28, 808–824.
- Ding, Y.; Shi, Y.; Yang, S. Molecular Regulation of Plant Responses to Environmental Temperatures. Mol. Plant 2020, 13, 544–564.
- Li, B.; Gao, K.; Ren, H.; Tang, W. Molecular Mechanisms Governing Plant Responses to High Temperatures. J. Integr. Plant Biol. 2018, 60, 757–779.
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Network of Plant Heat Stress Response. Trends Plant Sci. 2017, 22, 53–65.
- Gill, S.S.; Tuteja, N. Reactive Oxygen Species and Antioxidant Machinery in Abiotic Stress Tolerance in Crop Plants. Plant Physiol. Biochem. 2010, 48, 909–930.
- Wang, M.; Ding, F.; Zhang, S. Mutation of SlSBPASE Aggravates Chilling-Induced Oxidative Stress by Impairing Glutathione Biosynthesis and Suppressing Ascorbate-Glutathione Recycling in Tomato Plants. Front. Plant Sci. 2020, 11, 565701.
- Wang, M.; Zhang, S.; Ding, F. Melatonin Mitigates Chilling-Induced Oxidative Stress and Photosynthesis Inhibition in Tomato Plants. Antioxidants 2020, 9, 218.
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of Extreme Weather Disasters on Global Crop Production. Nature 2016, 529, 84–87.
- Asseng, S.; Ewert, F.; Martre, P.; Rötter, R.P.; Lobell, D.B.; Cammarano, D.; Kimball, B.A.; Ottman, M.J.; Wall, G.W.; White, J.W.; et al. Rising Temperatures Reduce Global Wheat Production. Nat. Clim. Chang. 2015, 5, 143–147.
- Wheeler, T.; Von Braun, J. Climate Change Impacts on Global Food Security. Science 2013, 341, 508–511.
- Lobell, D.B.; Schlenker, W.; Costa-Roberts, J. Climate Trends and Global Crop Production since 1980. Science 2011, 333, 616–620.
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P.; et al. Temperature Increase Reduces Global Yields of Major Crops in Four Independent Estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331.
- Zhang, J.; Li, X.M.; Lin, H.X.; Chong, K. Crop Improvement Through Temperature Resilience. Annu. Rev. Plant Biol. 2019, 70, 753–780.
- Aslam, M.; Fakher, B.; Ashraf, M.A.; Cheng, Y.; Wang, B.; Qin, Y. Plant Low-Temperature Stress: Signaling and Response. Agronomy 2022, 12, 702.
- Jung, J.H.; Seo, P.J.; Oh, E.; Kim, J. Temperature Perception by Plants. Trends Plant Sci. 2023, 28, 924–940.
- Andaya, V.C.; Mackill, D.J. Mapping of QTLs Associated with Cold Tolerance during the Vegetative Stage in Rice. J. Exp. Bot. 2003, 54, 2579–2585.
- Kagale, S.; Divi, U.K.; Krochko, J.E.; Keller, W.A.; Krishna, P. Brassinosteroid Confers Tolerance in Arabidopsis thaliana and Brassica napus to a Range of Abiotic Stresses. Planta 2007, 225, 353–364.
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate Action in Plant Growth and Development. J. Exp. Bot. 2017, 68, 1349–1359.
- Huang, X.; Shi, H.; Hu, Z.; Liu, A.; Amombo, E.; Chen, L.; Fu, J. ABA Is Involved in Regulation of Cold Stress Response in Bermudagrass. Front. Plant Sci. 2017, 8, 1613.
- Ding, F.; Ren, L.; Xie, F.; Wang, M.; Zhang, S. Jasmonate and Melatonin Act Synergistically to Potentiate Cold Tolerance in Tomato Plants. Front. Plant Sci. 2022, 12, 763284.
- Larkindale, J.; Knight, M.R. Protection against Heat Stress-Induced Oxidative Damage in Arabidopsis Involves Calcium, Abscisic Acid, Ethylene, and Salicylic Acid. Plant Physiol. 2002, 128, 682–695.
- Li, N.; Euring, D.; Cha, J.Y.; Lin, Z.; Lu, M.; Huang, L.J.; Kim, W.Y. Plant Hormone-Mediated Regulation of Heat Tolerance in Response to Global Climate Change. Front. Plant Sci. 2021, 11, 627969.
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, Perception, Signal Transduction and Action in Plant Stress Response, Growth and Development. An Update to the 2007 Review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058.
- Song, C.; Cao, Y.; Dai, J.; Li, G.; Manzoor, M.A.; Chen, C.; Deng, H. The Multifaceted Roles of MYC2 in Plants: Toward Transcriptional Reprogramming and Stress Tolerance by Jasmonate Signaling. Front. Plant Sci. 2022, 13, 868874.
- Wan, S.; Xin, X.F. Regulation and Integration of Plant Jasmonate Signaling: A Comparative View of Monocot and Dicot. J. Genet. Genom. 2022, 49, 704–714.
- Han, X.; Kui, M.; He, K.; Yang, M.; Du, J.; Jiang, Y.; Hu, Y. Jasmonate-Regulated Root Growth Inhibition and Root Hair Elongation. J. Exp. Bot. 2023, 74, 1176–1185.
- Huang, H.; Chen, Y.; Wang, S.; Qi, T.; Song, S. Jasmonate Action and Crosstalk in Flower Development and Fertility. J. Exp. Bot. 2023, 74, 1186–1197.
- Ding, F.; Wang, C.; Xu, N.; Zhang, S.; Wang, M. SlMYC2 Mediates Jasmonate-Induced Tomato Leaf Senescence by Promoting Chlorophyll Degradation and Repressing Carbon Fixation. Plant Physiol. Biochem. 2022, 180, 27–34.
- Ali, M.S.; Baek, K.H. Jasmonic Acid Signaling Pathway in Response to Abiotic Stresses in Plants. Int. J. Mol. Sci. 2020, 21, 621.
- Wang, F.; Guo, Z.; Li, H.; Wang, M.; Onac, E.; Zhou, J.; Xia, X.; Shi, K.; Yu, J.; Zhou, Y. Phytochrome A and B Function Antagonistically to Regulate Cold Tolerance via Abscisic Acid-Dependent Jasmonate Signaling. Plant Physiol. 2016, 170, 459–471.
- Liu, W.; Wang, H.; Chen, Y.; Zhu, S.; Chen, M.; Lan, X.; Chen, G.; Liao, Z. Cold Stress Improves the Production of Artemisinin Depending on the Increase in Endogenous Jasmonate. Biotechnol. Appl. Biochem. 2017, 64, 305–314.
- Seleiman, M.F.; Al-Suhaibani, N.; Ali, N.; Akmal, M.; Alotaibi, M.; Refay, Y.; Dindaroglu, T.; Abdul-Wajid, H.H.; Battaglia, M.L. Drought Stress Impacts on Plants and Different Approaches to Alleviate Its Adverse Effects. Plants 2021, 10, 259.
- Sharma, A.; Kumar, V.; Shahzad, B.; Ramakrishnan, M.; Singh Sidhu, G.P.; Bali, A.S.; Handa, N.; Kapoor, D.; Yadav, P.; Khanna, K.; et al. Photosynthetic Response of Plants Under Different Abiotic Stresses: A Review. J. Plant Growth Regul. 2020, 39, 509–531.
- Kim, J.M.; To, T.K.; Matsui, A.; Tanoi, K.; Kobayashi, N.I.; Matsuda, F.; Habu, Y.; Ogawa, D.; Sakamoto, T.; Matsunaga, S.; et al. Acetate-Mediated Novel Survival Strategy against Drought in Plants. Nat. Plants 2017, 3, 17097.
- Shahzad, A.N.; Pitann, B.; Ali, H.; Qayyum, M.F.; Fatima, A.; Bakhat, H.F. Maize Genotypes Differing in Salt Resistance Vary in Jasmonic Acid Accumulation During the First Phase of Salt Stress. J. Agron. Crop Sci. 2015, 201, 443–451.
- De Domenico, S.; Taurino, M.; Gallo, A.; Poltronieri, P.; Pastor, V.; Flors, V.; Santino, A. Oxylipin Dynamics in Medicago Truncatula in Response to Salt and Wounding Stresses. Physiol. Plant. 2019, 165, 198–208.
- Prerostova, S.; Dobrev, P.I.; Gaudinova, A.; Hosek, P.; Soudek, P.; Knirsch, V.; Vankova, R. Hormonal Dynamics during Salt Stress Responses of Salt-Sensitive Arabidopsis thaliana and Salt-Tolerant Thellungiella salsuginea. Plant Sci. 2017, 264, 188–198.
- Khan, A.H.; Ma, Y.; Wu, Y.; Akbar, A.; Shaban, M.; Ullah, A.; Deng, J.; Khan, A.S.; Chi, H.; Zhu, L.; et al. High-Temperature Stress Suppresses Allene Oxide Cyclase 2 and Causes Male Sterility in Cotton by Disrupting Jasmonic Acid Signaling. Crop J. 2023, 11, 33–45.
- Su, Y.; Huang, Y.; Dong, X.; Wang, R.; Tang, M.; Cai, J.; Chen, J.; Zhang, X.; Nie, G. Exogenous Methyl Jasmonate Improves Heat Tolerance of Perennial Ryegrass Through Alteration of Osmotic Adjustment, Antioxidant Defense, and Expression of Jasmonic Acid-Responsive Genes. Front. Plant Sci. 2021, 12, 664519.
- Li, M.; Yu, G.; Cao, C.; Liu, P. Metabolism, Signaling, and Transport of Jasmonates. Plant Commun. 2021, 2, 100231.
- Heitz, T.; Smirnova, E.; Widemann, E.; Aubert, Y.; Pinot, F.; Ménard, R. The Rise and Fall of Jasmonate Biological Activities. In Lipids in Plant and Algae Development; Subcellular Biochemistry; Springer: Cham, Switzerland, 2016; Volume 86.
- Koo, A.J. Metabolism of the Plant Hormone Jasmonate: A Sentinel for Tissue Damage and Master Regulator of Stress Response. Phytochem. Rev. 2018, 17, 51–80.
- Wasternack, C.; Feussner, I. The Oxylipin Pathways: Biochemistry and Function. Annu. Rev. Plant Biol. 2018, 69, 363–386.
- Kim, H.; Seomun, S.; Yoon, Y.; Jang, G. Jasmonic Acid in Plant Abiotic Stress Tolerance and Interaction with Abscisic Acid. Agronomy 2021, 11, 1886.
- Howe, G.A.; Major, I.T.; Koo, A.J. Modularity in Jasmonate Signaling for Multistress Resilience. Annu. Rev. Plant Biol. 2018, 69, 387–415.
- Chini, A.; Monte, I.; Zamarreño, A.M.; Hamberg, M.; Lassueur, S.; Reymond, P.; Weiss, S.; Stintzi, A.; Schaller, A.; Porzel, A.; et al. An OPR3-Independent Pathway Uses 4,5-Didehydrojasmonate for Jasmonate Synthesis. Nat. Chem. Biol. 2018, 14, 171–178.
- Schilmiller, A.L.; Koo, A.J.K.; Howe, G.A. Functional Diversification of Acyl-Coenzyme A Oxidases in Jasmonic Acid Biosynthesis and Action. Plant Physiol. 2007, 143, 812–824.
- Stenzel, I.; Hause, B.; Miersch, O.; Kurz, T.; Maucher, H.; Weichert, H.; Ziegler, J.; Feussner, I.; Wasternack, C. Jasmonate Biosynthesis and the Allene Oxide Cyclase Family of Arabidopsis thaliana. Plant Mol. Biol. 2003, 51, 895–911.
- Bannenberg, G.; Martínez, M.; Hamberg, M.; Castresana, C. Diversity of the Enzymatic Activity in the Lipoxygenase Gene Family of Arabidopsis thaliana. Lipids 2009, 44, 85–95.
- Grebner, W.; Stingl, N.E.; Oenel, A.; Mueller, M.J.; Berger, S. Lipoxygenase6-Dependent Oxylipin Synthesis in Roots Is Required for Abiotic and Biotic Stress Resistance of Arabidopsis. Plant Physiol. 2013, 161, 2159–2170.
- Chauvin, A.; Caldelari, D.; Wolfender, J.L.; Farmer, E.E. Four 13-Lipoxygenases Contribute to Rapid Jasmonate Synthesis in Wounded Arabidopsis thaliana Leaves: A Role for Lipoxygenase 6 in Responses to Long-Distance Wound Signals. New Phytol. 2013, 197, 566–575.
- Park, J.H.; Halitschke, R.; Kim, H.B.; Baldwin, I.T.; Feldmann, K.A.; Feyereisen, R. A Knock-out Mutation in Allene Oxide Synthase Results in Male Sterility and Defective Wound Signal Transduction in Arabidopsis Due to a Block in Jasmonic Acid Biosynthesis. Plant J. 2002, 31, 1–12.
- Stenzel, I.; Otto, M.; Delker, C.; Kirmse, N.; Schmidt, D.; Miersch, O.; Hause, B.; Wasternack, C. ALLENE OXIDE CYCLASE (AOC) Gene Family Members of Arabidopsis Thal Thaliana: Tissue-and Organ-Specific Promoter Activities and in Vivo Heteromerization. J. Exp. Bot. 2012, 63, 6125–6138.
- Theodoulou, F.L.; Job, K.; Slocombe, S.P.; Footitt, S.; Holdsworth, M.; Baker, A.; Larson, T.R.; Graham, I.A. Jasmonic Acid Levels Are Reduced in COMATOSE ATP-Binding Cassette Transporter Mutants. Implications for Transport of Jasmonate Precursors into Peroxisomes. Plant Physiol. 2005, 137, 835–840.
- Schneider, K.; Kienow, L.; Schmelzer, E.; Colby, T.; Bartsch, M.; Miersch, O.; Wasternack, C.; Kombrink, E.; Stuible, H.P. A New Type of Peroxisomal Acyl-Coenzyme a Synthetase from Arabidopsis thaliana Has the Catalytic Capacity to Activate Biosynthetic Precursors of Jasmonic Acid. J. Biol. Chem. 2005, 280, 13962–13972.
- Staswick, P.E.; Tiryaki, I. The Oxylipin Signal Jasmonic Acid Is Activated by an Enzyme That Conjugate It to Isoleucine in Arabidopsis W inside Box Sign. Plant Cell 2004, 16, 2117–2127.
- Li, C.; Schilmiller, A.L.; Liu, G.; Gyu, I.L.; Jayanty, S.; Sageman, C.; Vrebalov, J.; Giovannoni, J.J.; Yagi, K.; Kobayashi, Y.; et al. Role of β-Oxidation in Jasmonate Biosynthesis and Systemic Wound Signaling in Tomato. Plant Cell 2005, 17, 971–986.
- Suza, W.P.; Staswick, P.E. The Role of JAR1 in Jasmonoyl-l-Isoleucine Production during Arabidopsis Wound Response. Planta 2008, 227, 1221–1232.
- Caarls, L.; Elberse, J.; Awwanah, M.; Ludwig, N.R.; De Vries, M.; Zeilmaker, T.; Van Wees, S.C.M.; Schuurink, R.C.; Van den Ackerveken, G. Arabidopsis JASMONATE-INDUCED OXYGENASES down-Regulate Plant Immunity by Hydroxylation and Inactivation of the Hormone Jasmonic Acid. Proc. Natl. Acad. Sci. USA 2017, 114, 6388–6393.
- Smirnova, E.; Marquis, V.; Poirier, L.; Aubert, Y.; Zumsteg, J.; Ménard, R.; Miesch, L.; Heitz, T. Jasmonic Acid Oxidase 2 Hydroxylates Jasmonic Acid and Represses Basal Defense and Resistance Responses against Botrytis Cinerea Infection. Mol. Plant 2017, 10, 1159–1173.
- Feys, B.J.; Benedetti, C.E.; Penfold, C.N.; Turner, J.G. Arabidopsis Mutants Selected for Resistance to the Phytotoxin Coronatine Are Male Sterile, Insensitive to Methyl Jasmonate, and Resistant to a Bacterial Pathogen. Plant Cell 1994, 6, 751–759.
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-Iso-Jasmonoyl-L-Isoleucine Is the Endogenous Bioactive Jasmonate. Nat. Chem. Biol. 2009, 5, 344–350.
- Xie, D.; Xie, D.; Feys, B.F.; James, S.; Nieto-rostro, M.; Turner, J.G. COI1: An Arabidopsis Gene Required for Jasmonate-Regulated Defense and Fertility. Science 1998, 280, 1091–1994.
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ Family of Repressors Is the Missing Link in Jasmonate Signalling. Nature 2007, 448, 666–671.
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ Repressor Proteins Are Targets of the SCFCOI1 Complex during Jasmonate Signalling. Nature 2007, 448, 661–665.
- Yan, Y.; Stolz, S.; Chételat, A.; Reymond, P.; Pagni, M.; Dubugnon, L.; Farmer, E.E. A Downstream Mediator in the Growth Repression Limb of the Jasmonate Pathway. Plant Cell 2007, 19, 2470–2483.
- Chung, H.S.; Niu, Y.; Browse, J.; Howe, G.A. Top Hits in Contemporary JAZ: An Update on Jasmonate Signaling. Phytochemistry 2009, 70, 1547–1559.
- Vanholme, B.; Grunewald, W.; Bateman, A.; Kohchi, T.; Gheysen, G. The Tify Family Previously Known as ZIM. Trends Plant Sci. 2007, 12, 239–244.
- Nishii, A.; Takemura, M.; Fujita, H.; Shikata, M.; Yokota, A.; Kohchi, T. Characterization of a Novel Gene Encoding a Putative Single Zinc-Finger Protein, Zim, Expressed during the Reproductive Phase in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2000, 64, 1402–1409.
- Yan, J.; Zhang, C.; Gu, M.; Bai, Z.; Zhang, W.; Qi, T.; Cheng, Z.; Peng, W.; Luo, H.; Nan, F.; et al. The Arabidopsis CORONATINE INSENSITIVE1 Protein Is a Jasmonate Receptor. Plant Cell 2009, 21, 2220–2236.
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.-F.; Sharon, M.; Browse, J.; et al. Jasmonate Perception by Inositol Phosphate-Potentiated COI1- JAZ Co-Receptor. Nature 2010, 468, 400–405.
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, Metabolism, and Signaling by Proteins Activating and Repressing Transcription. J. Exp. Bot. 2017, 68, 1303–1321.
- Figueroa, P.; Browse, J. The Arabidopsis JAZ2 Promoter Contains a G-Box and Thymidine-Rich Module That Are Necessary and Sufficient for Jasmonate-Dependent Activation by MYC Transcription Factors and Repression by JAZ Proteins. Plant Cell Physiol. 2012, 53, 330–343.
- Liu, Y.; Du, M.; Deng, L.; Shen, J.; Fang, M.; Chen, Q.; Lu, Y.; Wang, Q.; Li, C.; Zhai, Q. Myc2 Regulates the Termination of Jasmonate Signaling via an Autoregulatory Negative Feedback Loop. Plant Cell 2019, 31, 106–127.
- Goossens, J.; Swinnen, G.; Vanden Bossche, R.; Pauwels, L.; Goossens, A. Change of a Conserved Amino Acid in the MYC2 and MYC3 Transcription Factors Leads to Release of JAZ Repression and Increased Activity. New Phytol. 2015, 206, 1229–1237.
- Dombrecht, B.; Gang, P.X.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 Differentially Modulates Diverse Jasmonate-Dependent Functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245.
- Fernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.-M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis BHLH Transcription Factors MYC3 and MYC4 Are Targets of JAZ Repressors and Act Additively with MYC2 in the Activation of Jasmonate Responses. Plant Cell 2011, 23, 701–715.
- Hu, Y.; Jiang, L.; Wang, F.; Yu, D. Jasmonate Regulates the INDUCER OF CBF EXPRESSION-C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 Cascade and Freezing Tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924.
- Ding, F.; Wang, C.; Xu, N.; Wang, M.; Zhang, S. Jasmonic Acid-Regulated Putrescine Biosynthesis Attenuates Cold-Induced Oxidative Stress in Tomato Plants. Sci. Hortic. 2021, 288, 110373.
- Ding, F.; Wang, C.; Zhang, S.; Wang, M. A Jasmonate-Responsive Glutathione S-Transferase Gene SlGSTU24 Mitigates Cold-Induced Oxidative Stress in Tomato Plants. Sci. Hortic. 2022, 303, 111231.
- Du, H.; Liu, H.; Xiong, L. Endogenous Auxin and Jasmonic Acid Levels Are Differentially Modulated by Abiotic Stresses in Rice. Front. Plant Sci. 2013, 4, 397.
- Habibi, F.; Ramezanian, A.; Rahemi, M.; Eshghi, S.; Guillén, F.; Serrano, M.; Valero, D. Postharvest Treatments with γ-Aminobutyric Acid, Methyl Jasmonate, or Methyl Salicylate Enhance Chilling Tolerance of Blood Orange Fruit at Prolonged Cold Storage. J. Sci. Food Agric. 2019, 99, 6408–6417.
- Zhao, M.L.; Wang, J.N.; Shan, W.; Fan, J.G.; Kuang, J.F.; Wu, K.Q.; Li, X.P.; Chen, W.X.; He, F.Y.; Chen, J.Y.; et al. Induction of Jasmonate Signalling Regulators MaMYC2s and Their Physical Interactions with MaICE1 in Methyl Jasmonate-Induced Chilling Tolerance in Banana Fruit. Plant Cell Environ. 2013, 36, 30–51.
- Jin, P.; Duan, Y.; Wang, L.; Wang, J.; Zheng, Y. Reducing Chilling Injury of Loquat Fruit by Combined Treatment with Hot Air and Methyl Jasmonate. Food Bioprocess Technol. 2014, 7, 2259–2266.
- González-Aguilar, G.A.; Tiznado-Hernández, M.E.; Zavaleta-Gatica, R.; Martínez-Téllez, M.A. Methyl Jasmonate Treatments Reduce Chilling Injury and Activate the Defense Response of Guava Fruits. Biochem. Biophys. Res. Commun. 2004, 313, 694–701.
- Jin, P.; Zheng, Y.; Tang, S.; Rui, H.; Wang, C.Y. A Combination of Hot Air and Methyl Jasmonate Vapor Treatment Alleviates Chilling Injury of Peach Fruit. Postharvest Biol. Technol. 2009, 52, 24–29.
- Ba, L.J.; Kuang, J.F.; Chen, J.Y.; Lu, W.J. MaJAZ1 Attenuates the MaLBD5-Mediated Transcriptional Activation of Jasmonate Biosynthesis Gene MaAOC2 in Regulating Cold Tolerance of Banana Fruit. J. Agric. Food Chem. 2016, 64, 738–745.
- Mao, D.; Xin, Y.; Tan, Y.; Hu, X.; Bai, J.; Liu, Z.Y.; Yu, Y.; Li, L.; Peng, C.; Fan, T.; et al. Natural Variation in the HAN1 Gene Confers Chilling Tolerance in Rice and Allowed Adaptation to a Temperate Climate. Proc. Natl. Acad. Sci. USA 2019, 116, 3494–3501.
- Zheng, Y.; Luo, L.; Wei, J.; Chen, Q.; Yang, Y.; Hu, X.; Kong, X. The Glutamate Receptors AtGLR1.2 and AtGLR1.3 Increase Cold Tolerance by Regulating Jasmonate Signaling in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2018, 506, 895–900.
- Ming, R.; Zhang, Y.; Wang, Y.; Khan, M.; Dahro, B.; Liu, J. The JA-responsive MYC2- BADH-like Transcriptional Regulatory Module in Poncirus Trifoliata Contributes to Cold Tolerance by Modulation of Glycine Betaine Biosynthesis. New Phytol. 2020, 229, 2730–2750.
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.-h.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A Regulator of Cold-Induced Transcriptome and Freezing Tolerance in Arabidopsis. Genes. Dev. 2003, 17, 1043–1054.
- Thomashow, M.F. Plant Cold Acclimation: Freezing Tolerance Genes and Regulatory Mechanisms. Annu. Rev. Plant Biol. 1999, 50, 571–599.
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 Encodes an AP2 Domain-Containing Transcriptional Activator That Binds to the C-Repeat/DRE, a Cis-Acting DNA Regulatory Element That Stimulates Transcription in Response to Low Temperature and Water Deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040.
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two Transcription Factors, DREB1 and DREB2, with an EREBP/AP2 DNA Binding Domain Separate Two Cellular Signal Transduction Pathways in Drought- and Low-Temperature-Responsive Gene Expression, Respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406.
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A Novel Cis-Acting Element in an Arabidopsis Gene Is Involved in Responsiveness to Drought, Low-Temperature, or High-Salt Stress. Plant Cell 1994, 6, 251.
- Shinwari, Z.K.; Nakashima, K.; Miura, S.; Kasuga, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. An Arabidopsis Gene Family Encoding DRE/CRT Binding Proteins Involved in Low-Temperature-Responsive Gene Expression. Biochem. Biophys. Res. Commun. 1998, 250, 161–170.
- Zhao, C.; Zhang, Z.; Xie, S.; Si, T.; Li, Y.; Zhu, J.K. Mutational Evidence for the Critical Role of CBF Transcription Factors in Cold Acclimation in Arabidopsis. Plant Physiol. 2016, 171, 2744–2759.
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The Cbfs Triple Mutants Reveal the Essential Functions of CBFs in Cold Acclimation and Allow the Definition of CBF Regulons in Arabidopsis. New Phytol. 2016, 212, 345–353.
- Thomashow, M.F. Molecular Basis of Plant Cold Acclimation: Insights Gained from Studying the CBF Cold Response Pathway. Plant Physiol. 2010, 154, 571–577.
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional Regulatory Networks in Cellular Responses and Tolerance to Dehydration and Cold Stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803.
- Kim, J.S.; Kidokoro, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA Demethylase ROS1 Prevents Inheritable DREB1A/CBF3 Repression by Transgene-Induced Promoter Methylation in the Arabidopsis Ice1-1 Mutant. Plant Mol. Biol. 2020, 104, 575–582.
- Thomashow, M.F.; Torii, K.U. Screaming Twist on the Role of ICE1 in Freezing Tolerance. Plant Cell 2020, 32, 816–819.
- Kidokoro, S.; Kim, J.S.; Ishikawa, T.; Suzuki, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DREB1A/CBF3 Is Repressed by Transgene-Induced DNA Methylation in the Arabidopsis Ice1-1 Mutant. Plant Cell 2020, 32, 1035–1048.
- An, J.; Wang, X.; Zhang, X.; You, C.; Hao, Y. Apple B-box Protein BBX37 Regulates Jasmonic Acid-mediated Cold Tolerance through the JAZ-BBX37-ICE1-CBF Pathway and Undergoes MIEL1-mediated Ubiquitination and Degradation. New Phytol. 2020, 229, 2707–2729.
- Boyce, J.M.; Knight, H.; Deyholos, M.; Openshaw, M.R.; Galbraith, D.W.; Warren, G.; Knight, M.R. The Sfr6 Mutant of Arabidopsis Is Defective in Transcriptional Activation via CBF/DREB1 and DREB2 and Shows Sensitivity to Osmotic Stress. Plant J. 2003, 34, 395–406.
- Knight, H.; Veale, E.L.; Warren, G.J.; Knight, M.R. The Sfr6 Mutation in Arabidopsis Suppresses Low-Temperature Induction of Genes Dependent on the CRT/DRE Sequence Motif. Plant Cell 1999, 11, 875.
- Knight, H.; Mugford, S.G.; Ülker, B.; Gao, D.; Thorlby, G.; Knight, M.R. Identification of SFR6, a Key Component in Cold Acclimation Acting Post-Translationally on CBF Function. Plant J. 2009, 58, 97–108.
- Zhang, X.; Wang, C.; Zhang, Y.; Sun, Y.; Mou, Z. The Arabidopsis Mediator Complex Subunit16 Positively Regulates Salicylate-Mediated Systemic Acquired Resistance and Jasmonate/Ethylene-Induced Defense Pathways. Plant Cell 2012, 24, 4294–4309.
- Wathugala, D.L.; Hemsley, P.A.; Moffat, C.S.; Cremelie, P.; Knight, M.R.; Knight, H. The Mediator Subunit SFR6/MED16 Controls Defence Gene Expression Mediated by Salicylic Acid and Jasmonate Responsive Pathways. New Phytol. 2012, 195, 217–230.
More
Information
Subjects:
Plant Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
979
Revisions:
2 times
(View History)
Update Date:
04 Mar 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




