Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Fei Ding and Version 2 by Rita Xu.
Temperature is a critical environmental factor that plays a vital role in plant growth and development. Temperatures below or above the optimum ranges lead to cold or heat stress, respectively. Temperature stress retards plant growth and development, and it reduces crop yields. Jasmonates (JAs) are a class of oxylipin phytohormones that play various roles in growth, development, and stress response.
- heat stress
- temperature stress
- jasmonates
1. Introduction
Plants are unable to move and thus have to cope with various adverse environmental factors, such as extreme temperatures, drought, salinity, and heavy metal toxicity [1][2][1,2]. Being a critical environmental factor, temperature plays a dominant role in plant growth and development [3], and it determines the geographical distribution of plant species [4]. As global climate change intensifies, the magnitude and frequency of extreme temperatures are increasing. Extreme temperatures cause various forms of damage at different stages during plant growth and development. The general consequences of heat and cold stress include impaired photosynthesis, excessive accumulation of reactive oxygen species, broken plasma membranes, and altered phytohormone levels [5][6][7][8][9][10][11][5,6,7,8,9,10,11]. Eventually, heat and cold stress inhibit plant growth and cause losses in crop yields, posing a serious threat to food security [12][13][14][15][12,13,14,15]. A survey showed that wheat, rice, maize, and soybean yields would decrease by 6.0%, 3.2%, 7.4%, and 3.1% on average, respectively, if the global temperature rises by 1 °C [4][16][4,16]. Low temperatures adversely affect plant growth and development and reduce crop production [12][17][18][19][12,17,18,19]. For instance, it has been estimated that in temperate and high-elevation regions, cold stress accounts for 30–40% of yield losses in rice [4][20][4,20].
Unlike animals, plants are sessile organisms and thus are unable to escape unfavorable temperature conditions. Instead, plants have evolved a set of sophisticated strategies enabling them to survive under temperature stress. Plant hormones play a vital role in the initiation of temperature stress response by integrating temperature stimulus and endogenous signals. For instance, jasmonates (JAs), abscisic acid (ABA), and brassinosteroid (BR) positively regulate plant response to both heat and cold stress [21][22][23][24][25][26][21,22,23,24,25,26]. Jasmonates (JAs) are a typical class of phytohormones. The term “jasmonates” generally refers to jasmonic acid and its derivatives, typically including jasmonyl isoleucine (JA-Ile) and methyl jasmonate (MeJA) [27]. In addition to its well-known role in growth and development, and in defense against pathogen attack and insect herbivory [28][29][30][31][32][28,29,30,31,32], a growing number of studies have highlighted the vital role of JA in the response to a variety of abiotic stresses, including drought, salinity, heat, and cold stress response [33][34][35][36][37][38][39][40][41][42][43][33,34,35,36,37,38,39,40,41,42,43].
2. JA Biosynthesis and Signaling
The JA biosynthetic pathway and the major enzymes involved have been well characterized and extensively reviewed [27][29][44][45][46][47][48][27,29,44,45,46,47,48]. JA biosynthesis starts with polyunsaturated fatty acids released from plastid membranes through the action of phospholipase (PLA) [49]. Current evidence supports the assertion that JA is derived via the α-linolenic acid (α-LeA, C18:3) pathway and the hexadecatrienoic acid (HTA, C16:3) pathway [50]. As the α-LeA pathway is dominant in the biosynthesis of JA, reswearchers focus on this pathway to explain JA biosynthesis (Figure 1A). Overall, four major enzymes are engaged in JA biosynthesis from α-LeA, comprising lipoxygenase (LOX), allene oxide synthase (AOS), allene oxide cyclase (AOC), and oxophytodienoic acid reductase (OPR) [51][52][51,52]. In plastids, LOX catalyzes the first step of JA biosynthesis. α-LeA is converted to 13(S)-hydroperoxy-octadecatrienoic acid (13-HPOT) by LOXs. In Arabidopsis, four LOXs—LOX2, LOX3, LOX4, and LOX6—are able to oxygenate α-LeA [53]. Each LOX may function differentially depending on the types of external stimuli. For instance, LOX6 is predominantly involved in JA production upon wounding and drought stress [54][55][54,55]. Subsequently, 13-HPOT is catalyzed by AOS to produce 12,13(S)-epoxy-octadecatrienoic acid (12,13-EOT). AOS is a cytochrome P450 enzyme, which uses oxygenated fatty acid hydroperoxide substrates as oxygen donors. There is only one AOS gene in Arabidopsis, and mutation in AOS leads to disrupted JA biosynthesis in response to wounding [56]. 12,13-EOT is further converted to 12-oxo-phytodienoic acid (12-OPDA), catalyzed by AOC. Four AOC genes have been identified and found to act redundantly in the biosynthesis of JA in Arabidopsis [57]. Next, 12-OPDA is translocated by the transporter COMATOSE (CTS1) to the peroxisome [58]. In the peroxisome, OPDA is reduced by OPDA reductase (OPR) to produce OPC-8:0. OPRs are encoded by six OPR genes in the Arabidopsis genome; however, only OPR3 acts on OPDA. OPC-8:0 is then subjected to three rounds of β-oxidation by acyl-CoA oxidase (ACX), L-3-KETOACYLCOA THIOLASE (KAT), and multifunctional protein (MFP) [59]. Finally, JasmonoylCoA, which is generated through β-oxidation reaction, can be further cleaved by THIOESTERASE (TE) to produce (+)-7-iso-JA, which is then transported to the cytoplasm. In the cytoplasm, various JA derivatives are formed, including methyl jasmonate (MeJA) and JA-isoleucine (JA-Ile) [60][61][60,61]. The conjugation of (+)-7-iso-JA with isoleucine produces JA-Ile, and the reaction is catalyzed by jasmonate-amido synthetase (JAR1). The jar1 mutant was identified as the first JA-insensitive mutant, and the JA-Ile level in mutant plants is severely reduced [62]. The methylation of JA forms MeJA, with catalysis by jasmonate methyl transferase (JMT). JA-Ile and MeJA are active forms of JA, and they can be converted to inactive 12-OH-JA by jasmonate-induced oxygenases (JO) and jasmonic acid oxidases (JAO) [63][64][63,64].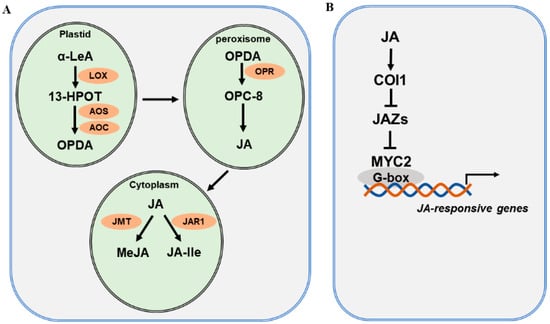
Figure 1. JA biosynthesis and signaling pathway. (A) A simplified JA biosynthesis pathway from α-linolenic acid (α-LeA). JA and its derivatives are produced from α-LeA through several sequential steps, which are catalyzed by lipoxygenase (LOX), allene oxide synthase (AOS), and allene oxide cyclase (AOC) in plastids; OPDA reductase (OPR) in peroxisomes; and jasmonate-amido synthetase (JAR1) and jasmonate methyl transferase (JMT) in the cytoplasm. (B) A simplified JA signaling pathway. Three main components are involved in JA signaling: the COI receptor, the JAZ repressor, and the MYC2 transcription factor.
