Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Roberto Piergentili | -- | 4905 | 2024-02-27 16:38:28 | | | |
| 2 | Catherine Yang | Meta information modification | 4905 | 2024-02-28 01:42:51 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Piergentili, R.; Marinelli, E.; Cucinella, G.; Lopez, A.; Napoletano, G.; Gullo, G.; Zaami, S. Epigenetics of BC and the Role of miR-125. Encyclopedia. Available online: https://encyclopedia.pub/entry/55560 (accessed on 07 February 2026).
Piergentili R, Marinelli E, Cucinella G, Lopez A, Napoletano G, Gullo G, et al. Epigenetics of BC and the Role of miR-125. Encyclopedia. Available at: https://encyclopedia.pub/entry/55560. Accessed February 07, 2026.
Piergentili, Roberto, Enrico Marinelli, Gaspare Cucinella, Alessandra Lopez, Gabriele Napoletano, Giuseppe Gullo, Simona Zaami. "Epigenetics of BC and the Role of miR-125" Encyclopedia, https://encyclopedia.pub/entry/55560 (accessed February 07, 2026).
Piergentili, R., Marinelli, E., Cucinella, G., Lopez, A., Napoletano, G., Gullo, G., & Zaami, S. (2024, February 27). Epigenetics of BC and the Role of miR-125. In Encyclopedia. https://encyclopedia.pub/entry/55560
Piergentili, Roberto, et al. "Epigenetics of BC and the Role of miR-125." Encyclopedia. Web. 27 February, 2024.
Copy Citation
Breast Cancer (BC) is one of the most common cancer types worldwide, and it is characterized by a complex etiopathogenesis, resulting in an equally complex classification of subtypes. MicroRNA (miRNA or miR) are small non-coding RNA molecules that have an essential role in gene expression and are significantly linked to tumor development and angiogenesis in different types of cancer. miR-125 is a highly conserved family of microRNAs whose members have also been found in nematodes (named lin-4 in 1993, the first miR described ever).
breast cancer (BC)
MicroRNA (miR)
non-coding RNA
competing endogenous RNA (ceRNA)
1. microRNA Nomenclature
MicroRNAs are short (20–25 nucleotides), single-stranded, non-coding RNA molecules whose main function is gene expression control, mainly silencing. They exert this downregulation by binding the 3′ end of target mRNA(s) through sequence homology and promoting either their degradation or impairing their translation [1]. Over 2500 miR have been estimated to be encoded in the human genome, regulating over 60% of human genes [2]. In addition, thanks to imperfect sequence pairing, they can also bind multiple targets, thus amplifying their intracellular action.
Beyond the number identifier (ID, usually higher for miR described chronologically later), additional nomenclature rules are established to identify unequivocally each miR [3][4]. miRs with almost identical sequence are identified by a progressive lowercase letter after the identification number (miR-XXXa, miR-XXXb, etc.), while miRs with identical sequence but mapping to different genomic locations are indicated by a progressive number separated from the ID number by a dash (e.g., miR-XXX-1, miR-XXX-2, etc.). To further distinguish molecules of different species, an additional three-letter code and a dash may be added at the beginning of the miR name (i.e., hsa-miR-XXX indicates a human–Homo sapiens–miR). Finally, the mature, single-stranded miR can be obtained either from the 5′ end or the 3′ end of its double-stranded miR precursor (pre-miR). Notably, sometimes both strands can—separately—be used for mRNA regulation, resulting in 5p and 3p miR forms if both are present and functional in the cell and the two miR are roughly equivalent in their intracellular amount; instead, if both are present but one is significantly more abundant than the other, then the rarer one has an asterisk at the end of its name (e.g., miR-XXX-5p*).
2. Role of miR in BC
Increasing evidence shows that miRs represent a central hub of gene expression control in human carcinogenesis, and, from this perspective, BC is not an exception [5][6].
Significantly, miR-21 has been shown to be responsible for the development of multidrug resistance [7], and it modulates the resistance of BC cells to doxorubicin by targeting PTEN [8]. In addition, miR-21 also plays a central role in BC proliferation and metastasis by targeting LZTFL1 [9]. Additional miR-21 targets involved in cell proliferation, metastasis, epithelial-to-mesenchymal transition (EMT), and apoptosis in BC include IGFBP3, TPM1, PCD4, and TGF-beta1 [10].
Another player in BC etiopathogenesis is miR-106a, which promotes cancer progression through the downregulation of RAF-1 [11], P53, BAX, and RUNX3 and the upregulation of Bcl-2 and ABCG2; it also confers cisplatin resistance upon its upregulation [12][13].
Upregulation of miR-155 causes telomere fragility through its action on TRF1, a component of the shelterin complex [14]. Interestingly, this miR, together with miR-10b, miR-34a and miR-141, is also a possible candidate for building a panel of circulating miR useful for non-invasive detection of this tumor [15].
Conversely, downregulated miR-141 has been reported to be a typical feature of BC, where its target is ANP32E [16], which, in turn, induces tumorigenesis in triple-negative BC (TNBC) cells by upregulating E2F1 [17]. Instead, high miR-141-3p expression is typical of grade III BC compared to grade II and, together with miR-181b1-5p and miR-23b-3p, it is a useful marker not only to discriminate between malignant and benign breast tissues but might also help in distinguishing TNBC from other molecular subtypes of BC [18].
The let-7 family of miR are tumor suppressors in several cancers, including BC, and their members can also be detected as circulating biomarkers [19]. Let-7 action involves the control of ERCC6 expression [20], and its overexpression could inhibit BC cell proliferation.
Another circulating marker of BC is miR-335 [21], which exerts its effects by simultaneously regulating the known BRCA1 activators ERα, IGF1R, SP1 and the repressor ID4, including a feedback regulation of miR-335 expression by estrogens [22]. Its overexpression causes decreased cell viability and increased apoptosis, while other findings show it to negatively regulate the HGF/c-Met pathway, thus affecting cell scattering, migration, and invasion [23].
Another downregulated miR in BC is miR-126 [24]. Its targets include VEGFA and PIK3R2 [25]. It also reduces trastuzumab resistance by targeting PIK3R2 and regulating the AKT/mTOR signaling pathway [26] and controls cell invasion by targeting ADAM9 [27].
Tumor suppressor miR-199a/b-3p inhibits migration and invasion of BC cells by downregulating the PAK4/MEK/ERK signaling pathway [28]. Overexpression of miR-199a-3p targets the c-Met and mTOR pathways, increases doxorubicin sensitivity and causes G1 phase arrest, thus reducing cell invasion and promoting doxorubicin-induced apoptosis [29]. This miR also confers resistance to cisplatin treatment by downregulating TFAM [30] and, at the same time, promotes BC development and metastasis under hypoxic conditions by controlling the regulatory axis consisting of HIF-1, SNHG1, and TFAM [31]. The same study mentioned above [29] also shows that in TNBC patients, additional circulating miR (i.e., miR-19a/b-3p, miR-25-3p, miR-22-3p, miR-210-3p and miR-93-5p) are deregulated as well and control several molecular pathways involved in drug resistance, making them amenable to be used as BC biomarkers, together with let-7a-5p, miR-100-5p and miR-101-3p, identified in another study [32].
Tumor suppressor miR-101 is downregulated as well in BC, and its targets include POMP, Stmn1, DNMT3A, EYA1, VHL, SOX2, Jak2 and MCL-1 (reviewed in [33]). For this reason, it plays a major role in the control of several cancer-related cellular processes, such as proliferation, apoptosis, angiogenesis, drug resistance, invasion, and metastasis. Overexpression of miR-101-3p can inhibit the migration of BC cells into the brain endothelium, a frequent and late event in BC patients, by inducing COX-2/MMP1 signaling, which can degrade the inter-endothelial junctions (claudin-5 and VE-cadherin) [34]. Jiang and collaborators showed that the suppression of the oncogene EZH2 in BC by miR-101-3p is potentiated in the presence of syn-cal14.1a, a synthetic peptide derived from Californiconus californicus (a sea snail), thus inhibiting cell migration, invasion, and proliferation [35]. Additional data come from the work of Toda and collaborators, who performed an RNA-sequence-based microRNA expression signature in BC and identified other dysregulated miRs in BC (e.g., miR-99a-5p/-3p, miR-101-5p/-3p, miR-126-5p/-3p, miR-143-5p/-3p, and miR-144-5p/-3p) and found that miR-101-5p controls the expression of seven putative oncogenes (i.e., HMGB3, ESRP1, GINS1, TPD52, SRPK1, VANGL1 and MAGOHB) [36].
Finally, miR-9 is known to exert critical functions in the initiation and progression of BC. Its upregulation—together with that of miR-221/222, miR-373 and miR-10b—is linked to highly malignant invasive EMT and cancer stem cell production [37]. Conversely, its downregulation can lead to improved overall survival, smaller tumors, earlier stages, and ER-positive cancers due to the enrichment of estrogen response genes [38]. Gwak and collaborators showed that miR-9 is highly expressed in HER2+ and TNBC subtypes compared with luminal subtypes, tumors with a high tumor stage or histologic grade, and tumors displaying the CD44+/CD24− phenotype, vimentin expression, and E-cadherin loss [39]. Interestingly, Shen and collaborators showed miR-9-5p, together with miR-195-5p and miR-203a-3p, to be a part of the extracellular vesicle (EV)-encapsulated miR (enabling cancer cell–cell communication in tumor pathogenesis and response to therapies) excreted upon docetaxel treatment [40]. As for its targets, genes identified so far include FOXO1 [41], STARD13 [42], LIFR [43], elf5A2 [44], HMGA2, EGR1, and IGFBP3 [10] and PDGFRbeta [45]. In turn, its expression is activated by MYC and MYCN [46][47].
A summary of the miR involved in BC and their function is summarized in Table 1.
Table 1. List of miR playing a direct role in BC. Target genes are those for which the miR/mRNA interaction is direct (usually, at the mRNA 3′ UTR), thus indirect interactions (e.g., other proteins of the same metabolic axis) are not reported in this table; additional miR studied only as BC biomarkers are collectively reported in the bottom row; see text for additional explanations. Abbreviations: n/a—data not available.
| miR Name | Target Gene(s) | Affected Cellular Functions | Refs |
|---|---|---|---|
| miR-21 | PTEN | drug resistance | [7][8] |
| miR-21 | LZTFL1 | proliferation and metastasis | [9] |
| miR-21 | IGFBP3 TPM1 PCD4 TGF-β1 |
proliferation, metastasis, epithelial-to-mesenchymal transition (EMT), apoptosis | [10] |
| miR-106a | RAF-1 | invasion and proliferation | [11] |
| miR-106a | P53 BAX RUNX3 Bcl-2 ABCG2 |
proliferation, colony-forming capacity, migration, invasion, apoptosis, sensitivity to cisplatin | [12][13] |
| miR-155 | TRF1 | telomere fragility | [14] |
| miR-141 | ANP32E | migration and invasion | [16] |
| let-7 | ERCC6 | proliferation, apoptosis | [20] |
| miR-335 | ERα IGF1R SP1 ID4 |
proliferation, apoptosis | [22] |
| miR-335 | c-Met | cell scattering, migration, and invasion | [23] |
| miR-126 | VEGFA PIK3R2 |
angiogenesis, tumor genesis and growth | [25] |
| miR-126 | PIK3R2 | trastuzumab resistance | [26] |
| miR-199a/b-3p | PAK4 | migration and invasion | [28] |
| miR-199a-3p | mTOR c-Met |
cell cycle progression, doxorubicin sensitivity, apoptosis | [29] |
| miR-199a-3p | TFAM | resistance to cisplatin | [30] |
| miR-199a-3p | TFAM | angiogenesis and metastasis under hypoxia | [31] |
| miR-101 | POMP Stmn1 DNMT3A EYA1 VHL SOX2 Jak2 MCL-1 |
proliferation, apoptosis, angiogenesis, drug resistance, invasion, metastasis | [33] |
| miR-101-3p | COX-2 | migration, metastasis | [34] |
| miR-101-3p | EZH2 | migration, invasion, proliferation | [35] |
| miR-101-5p | GINS1 | DNA replication | [36] |
| miR-9 | FOXO1 | proliferation, migration, invasion | [41] |
| miR-9 | STARD13 | EMT, metastasis | [42] |
| miR-9 | LIFR | metastasis | [43] |
| miR-9 | elf5A2 | resistance to doxorubicin | [44] |
| miR-9 | HMGA2 EGR1 IGFBP3 |
proliferation, metastasis, EMT, apoptosis | [10] |
| miR-9 | PDGFRβ | vasculogenesis | [45] |
| miR-200 | PDGFRβ | vasculogenesis | [45] |
| let-7a-5p miR-9-5p miR-10b miR-21 miR-22-3p miR-23b-3p miR-25-3p miR-29 miR-34a miR-93-5p miR-99a-5p/-3p miR-100-5p miR-101-3p miR-101-5p miR-126-5p/-3p miR-141-3p miR-143-5p/-3p miR-144-5p/-3p miR-145 miR-155 mir-181b1-5p miR-195-5p miR-199a-5p miR-200a miR-203 miR-203a-3p miR-205 miR-210-3p miR-221/222 miR-373 |
n/a | biomarkers | [15][18][29][32][36][37][39][40] |
All together, these data point to the involvement of several miRs in BC formation, development, metastasis, and drug resistance, showing that at the molecular level it is crucial to identify which pathways are altered and why, for example, the same gene may be deregulated because of the alteration of diverse miR. Knowing which miR is altered may greatly affect therapeutic approaches, especially in terms of avoiding cross-effects due to off-target actions. Thus, there is a necessity to identify (hopefully, all) the players in BC pathogenesis in a patient-specific way. In this perspective, the miR-125 family of miR has gained increasing relevance and attention in BC research, thanks to the numerous publications released over the last few years. In light of the numerous targets of these miRs and the multitude of pathways potentially altered inside the cell upon their dysregulation, in the next few years, miR-125 is likely to become central to understanding BC biology.
3. The miR-125 Family: Molecular Organization and Roles in Human Pathology
miR-125 is a highly conserved family of microRNAs whose members have also been found in nematodes (named lin-4 in 1993, the first miR described ever) [48]. The miR-125 family in H. sapiens includes three members, namely miR-125a, miR-125b-1 and miR-125b-2. The MIR125A gene maps to chromosome 19q13.41 [49], and miR-125a is part of a transcribed cluster of miR, together with miR-99b and let-7e [50]. The MIR125B1 gene maps to chromosome 11q24.1, and in this locus, it is part of a cluster including the LET7A2 and MIR100 genes [50][51]. These miRs are inside the third intron of the MIR100HG gene [52]. Finally, the MIR125B2 gene maps to chromosome 21q21.1, where it is included in a cluster together with the MIR99A and LET7C genes [50][51], inside the sixth intron of the MIR99AHG gene [52]. miR-125a and miR-125b differ only by a central diuridine insertion and a U-to-C change in miR-125a [53]. All members of the family show both 5p and 3p forms (Figure 1).
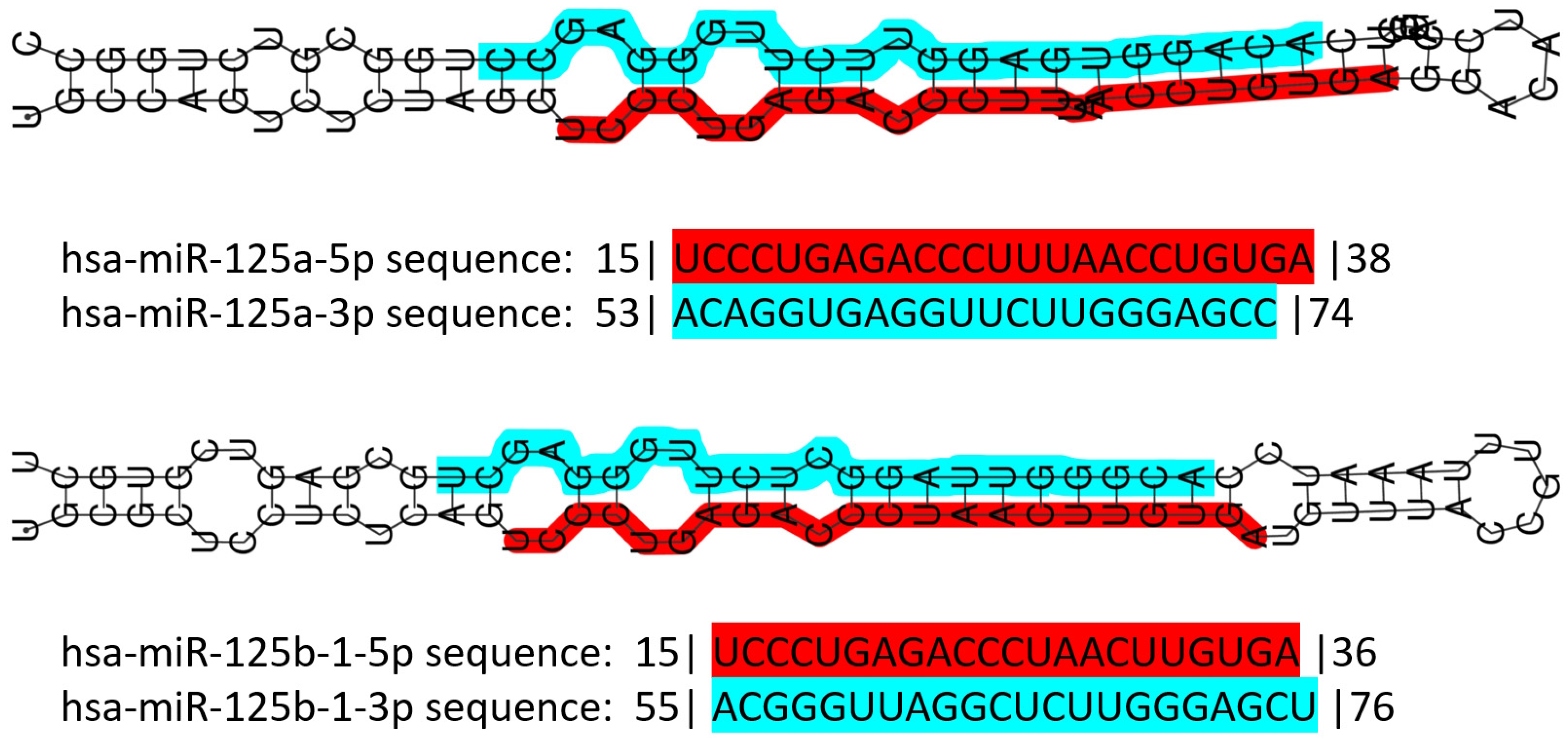
Figure 1. Schematic representation of human miR-125 illustrating the structure of the double-stranded pre-miR of both miR-125a and miR-125b, and their sequence. Color codes: red highlight for the 5p form, blue highlight for the 3p form. Numbers before and after the sequences indicate the number of nucleotides trimmed away from the mature miR. Data retrieved and partially modified from miRTarBase v9 update 2022 [54][55].
The miR-125 family is involved in several cell metabolic pathways controlling differentiation, proliferation, apoptosis, metastasis formation, drug resistance and immune system function because of the targeting of mRNAs related to these cellular processes [56] (Figure 2). miR-125 molecules have a complex behavior inside the cell, which mirrors their expression pattern in different tissues/cell types [54][55], their ample variety of targets [54][55], the intracellular role of their targets, and the way miR and mRNAs are either up- or down-regulated upon expression.
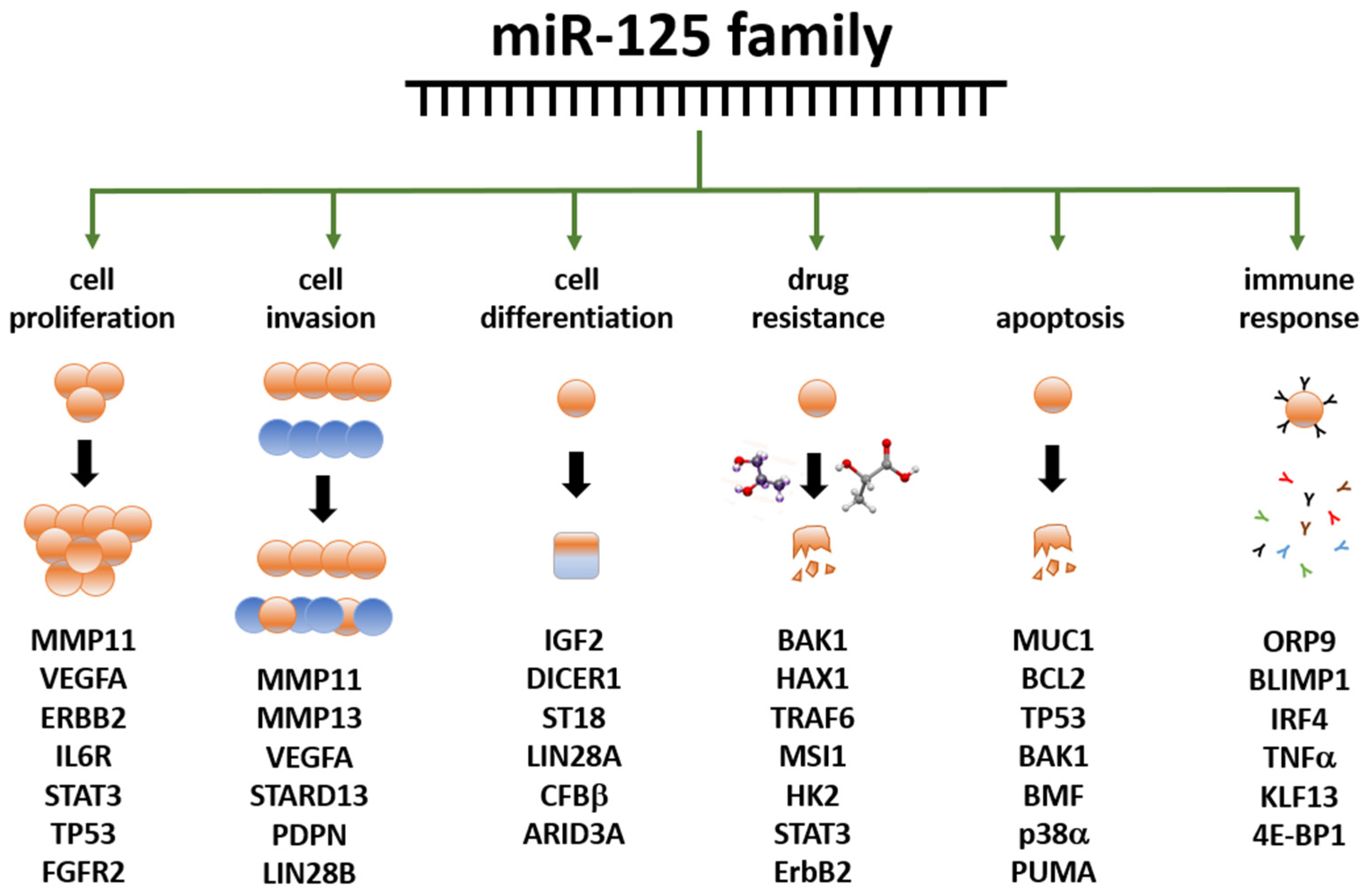
Figure 2. Summary of the main functions exerted by the miR-125 family members in human biology. Examples of miR-125 target genes are reported below each function. Data partly retrieved from [56][57] The listed genes at the bottom are involved in the control of the cellular functions below the green arrows; they are controlled, either directly or indirectly, by miR-125 family members, either by up- or down-regulation. Further details have been laid out in the article’s body.
The role of miR-125 family members has been extensively demonstrated in the muscle. It interacts with insulin-like growth factor II (IGF-II) to regulate myoblast differentiation in vitro and muscle regeneration in vivo [58], and with TRAF6 to prevent atrophy [59]. It is also involved in the proliferation and migration of vascular smooth muscle cells induced by platelet-derived growth factor BB [60]. In cardiac muscles, miR-125 participates in the development of the heart in embryonic mammals (reviewed in [61]); it regulates muscle-enriched transcription factors in cardiac and skeletal myocytes [62]; it can modulate cardiac progenitor cell proliferation and migration potential [63]; and it regulates cardiomyocytes proliferation and apoptosis under oxidative stress conditions [64]. Cardiac-specific miR-125b deficiency has recently been shown to induce perinatal death and cardiac hypertrophy [65].
miR-125 is one of the most abundant microRNAs in the central nervous system (CNS) in both mice and men [66]. In humans, miR-125b promotes neuronal differentiation in human cells by repressing at least ten target mRNAs involved in those pathways [67][68]. It also regulates dendritic spine morphology and synaptic maturation [69], it is implicated in synaptic plasticity [70], promotes astrogliogenesis, and is involved in astrogliosis and glial cell proliferation [71]. Its deregulation has also been linked to CNS tumor formation and growth, such as pediatric low-grade glioma [72]; it regulates cell growth arrest and apoptosis of human neuroblastoma- and medulloblastoma-derived cell lines [73][74]; it inhibits cell apoptosis through p53 and p38MAPK-independent pathways in glioblastoma cells [75]; and, in glioma, it targets BMF [76].
In the immune system, miR-125 regulates hematopoiesis, inflammation, and immune cell function. miR-125a controls stem cell homeostasis during hematopoiesis [77][78][79][80] and plays a role in immune cell identity [80]. miR-125-5p targeting IL-6R regulates macrophage inflammatory response and intestinal epithelial cell apoptosis in ulcerative colitis through the JAK1/STAT3 and NF-κB pathways [81]. miR-125b-1-3p is expressed in hMSCs-Ad exosomes and can promote T lymphocyte apoptosis and alleviate atherosclerosis (AS) by down-regulating BCL11B expression, thus providing potential molecular targets for the clinical treatment of AS [82].
All together, these data emphasize the multiple roles of miR-125 family members in cell proliferation and differentiation in numerous body locations.
4. miR-125 and Cancer
Studying the role of miR-125 in cancer is an important research area; beyond the above-mentioned tumors of the CNS, this noncoding RNA is indeed deregulated in several other tumors [83]. Additional organs affected by miR-125-related cancers include the ovary, bladder, liver, skin, bone, lung, pancreas, prostate, thyroid, stomach, colon and kidney. A summary of the cancers affecting human organs, known miR-125 targets and related bibliographic references are reported in Table 2.
Table 2. Summary of the affected organs and mRNA targets of miR-125 family members in human cancers, except for BC reported in the next section. Abbreviations: CNS—central nervous system; refs—references; n/a—data not available in the cited reference(s).
| miR | Organ | Target(s) | Notes | Refs |
|---|---|---|---|---|
| 125 | CNS | n/a | deregulated, pediatric | [72] |
| 125 | CNS | n/a | deregulated | [73][74] |
| 125 | CNS | p53, p38MAPK | none | [75] |
| 125 | CNS | BMF | none | [76] |
| 125a | ovary | n/a | EMT negative regulator | [84] |
| 125b | ovary | BCL3 | none | [85] |
| 125b | ovary | n/a | serum biomarker | [86] |
| 125b | bladder | E2F3 | none | [87] |
| 125b | bladder | n/a | urine biomarker | [88] |
| 125-3p | bladder | n/a | hypoxia regulated | [89] |
| 125 | bladder | n/a | survival predictor | [90] |
| 125a | liver | MMP11, VEGF | none | [91] |
| 125b | liver | Mcl-1, IL6R | none | [92] |
| 125b | liver | Lin28B2 | none | [93] |
| 125 | liver | Pokemon | none | [94] |
| 125 | liver | TRAF6 | none | [95] |
| 125 | liver | hexokinase II | none | [96] |
| 125 | liver | FOXM1 | none | [97] |
| 125 | skin | NCAM | none | [98] |
| 125 | skin | c-Jun | none | [99] |
| 125b | skin | MMP13 | none | [100] |
| 125b | skin | STAT3 | none | [101] |
| 125 | skin | n/a | deregulated | [102] |
| 125b | bone | STAT3 | none | [103][104] |
| 125 | bone | ErbB2 | none | [105] |
| 125 | bone | BAP1 | none | [106] |
| 125 | lung | n/a | survival predictor | [107] |
| 125 | lung | EGFR | none | [108] |
| 125 | lung | HER2 | trastuzumab resistance | [109] |
| 125 | lung | MMP13 | none | [110] |
| 125 | pancreas | n/a | deregulated | [111][112] |
| 125 | pancreas | NEDD9 | none | [113] |
| 125 | prostate | n/a | deregulated | [114][115][116] |
| 125 | prostate | BAK1 | none | [117] |
| 125 | prostate | p53, PUMA | none | [118] |
| 125b | thyroid | Foxp3 | cisplatin sensitivity | [119] |
| 125b | stomach | PPP1CA-Rb | none | [120] |
| 125a-5p | colon | BCL2, BCL2L12, MCL1 | none | [121] |
| 125b | kidney | n/a | survival predictor | [122] |
5. Role of miR-125 in BC
A relatively small amount of research is currently available on the role of miR-125 in BC. The reports showing its altered expression in these malignancies started to be published more than 20 years ago, and the research is still running in search of an affordable diagnostic panel based on this noncoding RNA [123][124][125][126][127][128]. Among the targets first identified, it is worth mentioning ERBB2 and ERBB3 [129] mRNA. In 2011, Zhang and colleagues demonstrated the action of miR-125b on the regulation of the ETS1 proto-oncogene in human invasive BC [130]. Rajabi et al. found that miR-125b, downregulated in BC, can reduce the expression of MUC1 (an oncoprotein), whose silencing causes DNA damage-induced apoptosis in cancer cells [131]. Tang and collaborators studied the effects of miR-125 deregulation on metastasis formation, finding that miR-125b induces metastasis by targeting STARD13 mRNA in MCF-7 and MDA-MB-231 BC cells [132], in contrast with the tumor suppressive action described before. Using the same BC cell lines, Metheetrairut and collaborators showed that forced expression of miR-125b results in radiosensitivity, as seen by reduced clonogenic survival, enhanced apoptotic activity and enhanced senescence post-ionizing radiation treatment. Moreover, re-expression of c-JUN in MDA-MB-231 cells promoted radioresistance and abrogated miR-125-mediated radiosensitization, suggesting that overexpression of miR-125b causes sensibilization to γ-irradiation and indicating this miR as a possible target for adjuvant therapy [133]. In contrast, Wang et al. found an association between miR-125b expression and chemoresistance [134], again indicating an oncogenic role for this miR. In line with these last results, Zhou and collaborators found that miR-125b confers the resistance of BC cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression [135]. He and collaborators studied the expression of miR-125a-5p/3p and miR-125b in 143 pairs of BC and normal adjacent tissues, finding that miR-125a-5p and miR-125b were significantly down-regulated in BC tissue samples and that the expression level of miR-125a-5p was significantly higher in younger patients (<35 years) than in older ones, and a gradual reduction in miR-125a-5p expression was observed in BC tissue samples correlated to increasing age [136]. Recently, a paper showed the oncosuppressor role of miR-125b via the inhibition of proliferation, migration, and invasion of BC cells through targeting MMP11 protein expression [137]. A summary of the data reported above is illustrated in Table 3.
Table 3. Role of miR-125 in BC formation and development. In columns 2 and 4, reg. stands for regulation; an arrow pointing upwards means upregulation, while an arrow pointing downwards means downregulation; each arrow describes the miR/target regulation reported to its left. Note that in BC models, miR-125 members may be either up- or down-regulated, indicating either an oncogenic or oncosuppressive role for this molecule, in that context. Possible interpretations of these contradictory data are reported in the Discussion. In columns 3–4, n/a stands for ‘data not available’ or ‘not applicable’. In column 6, ‘blood samples’ means that circulating miR have been studied. In column 7, ref. stands for reference(s).
| miR | Reg. | Target | Reg. | Cellular Function | Cell Line | Ref. |
|---|---|---|---|---|---|---|
| miR-125a miR-125b |
↑ ↑ |
ERBB2 ERBB3 |
↓ ↓ |
migration invasion |
SKBR3 | [129] |
| miR-125b | ↓ | ETS1 | ↑ | proliferation | BC samples | [130] |
| miR-125b | ↓ | MUC1 | ↑ | apoptosis | BT-549 ZR-75-1 |
[131] |
| miR-125b | ↓ | STARD13 | ↑ | metastasis | MCF-7 MDA-MB-231 |
[132] |
| miR-125 | ↓ | n/a | n/a | radioresistance | MCF-7 MDA-MB-231 |
[133] |
| miR-125b | ↑ | n/a | n/a | chemoresistance proliferation apoptosis |
blood samples | [134] |
| miR-125b | ↑ | BAK1 | ↓ | chemoresistance apoptosis |
MDA-435 MDA-436 MDA-231 MCF7 SKBR3 |
[135] |
| miR-125a-5p miR-125b |
↓ ↓ |
n/a | n/a | age-dependent BC formation | BC samples | [136] |
| miR-125b | ↓ | MMP11 | ↑ | proliferation migration invasion |
T47D SKBR3 |
[137] |
6. Further Mining miR-125 Function in BC: Competing Endogenous RNA Networks (ceRNET)
A fundamental way to control gene expression through miRs has been elucidated in recent years, consisting of the so-called ceRNET. In fact, miRs have been shown to act as controllers of target mRNAs by altering their half-life or translation. However, they are also controlled, in many cases, by other long non-coding RNAs (lncRNA) or even other mRNA, which “sponge” miR through sequence homology, avoiding their interaction with mRNA targets [138]. In other words, lncRNA and mRNA compete for binding miR; these two molecules form a competing endogenous RNA (ceRNA) couple. If the lncRNA efficiently sponges the miR, then miR inhibitory action is not accomplished, and the target mRNA is regularly translated. In this case, the lncRNA, inhibitor of an inhibitor, has a function resembling that of an enhancer of gene expression. Hence, if the mRNA encodes an oncoprotein, the lncRNA has an oncogenic effect, while the miR has an oncosuppressive role. The same, with opposite effects, occurs in the case of the mRNA coding an oncosuppressor. The three molecules, taken together, form what is currently known as a regulatory axis, and the sum of many axes creates the ceRNET. Here, lncRNA and mRNA constitute the nodes of the network, while miR represent their connections. A growing number of research works have been published in recent years outlining the increasing structure and complexity of the ceRNET in BC (see [139] and references therein), including the action of pseudogenes in this phenomenon. In fact, Welch and collaborators found that 309 pseudogenes exhibit significant differential expression among BC subtypes, and their expression pattern allows recognizing tumor samples from normal samples and discriminating the basal subtype from the luminal and Her2 subtypes; of them, 177 transcribed pseudogenes possess binding sites for co-expressed miRs that are also predicted to target their parent genes [140]. Recently, in a work by Zhu and collaborators, the authors took advantage of the data available in the exoRbase database and derived it from the exosomes of human BC samples [141]. Their study allowed for the identification of a ceRNA network including 19 mRNA nodes, 2 lncRNA nodes, 8 circular RNA nodes, and 41 miR connections. KEGG enrichment analysis showed that differentially expressed mRNA in the regulatory network is mainly enriched in the p53 signaling pathway.
Research about portions of a miR-125-centered ceRNET has been expanding steadily. The miR-125 interactions with the mRNA described in the previous section are therefore likely to become axes of the growing BC ceRNET as well, as soon as the appropriate lncRNA is identified in the pathway. However, some axes have already been described, and some of them, being interconnected, can be used to build a basic version of this network (Figure 3).
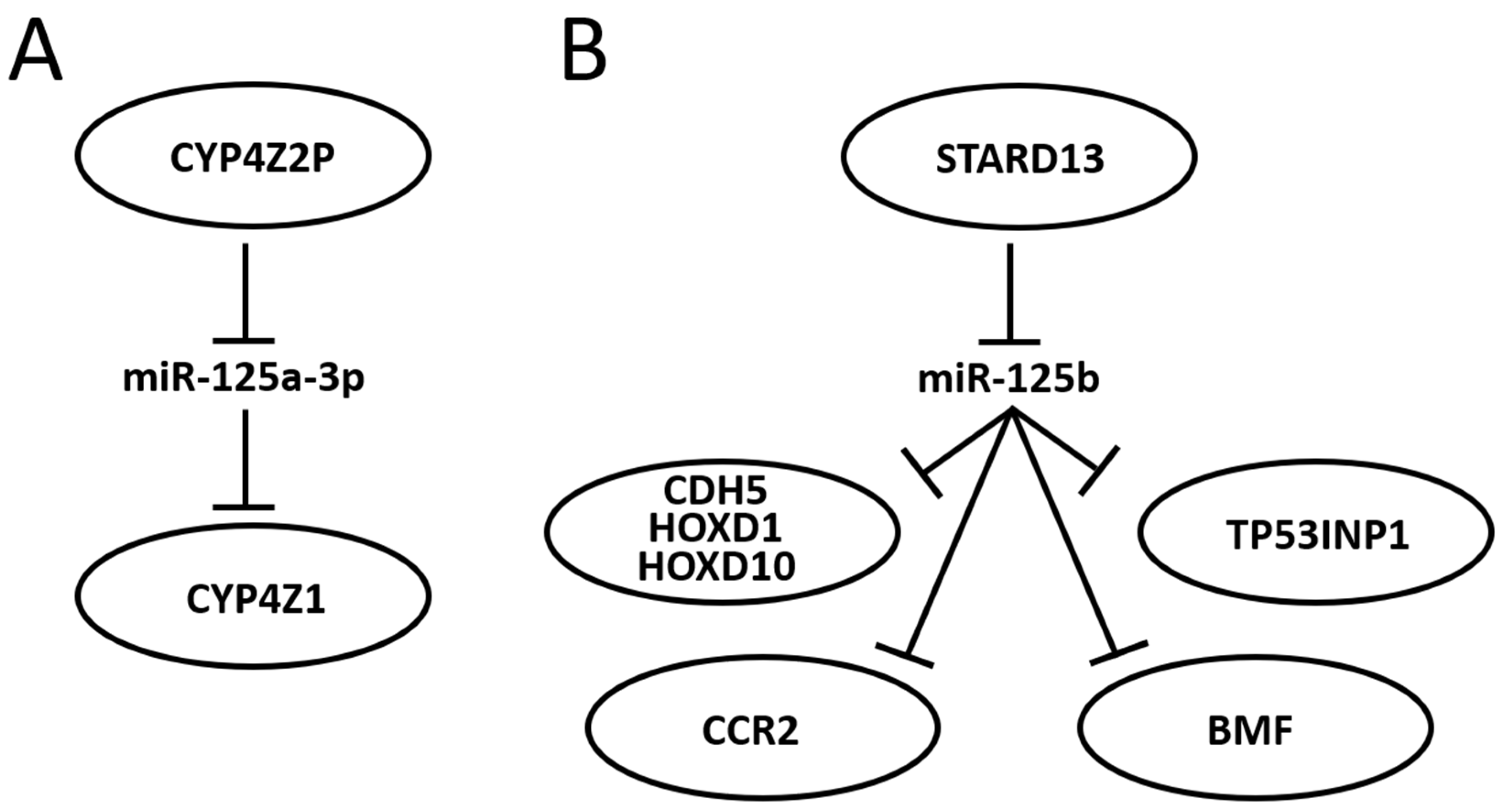
Figure 3. Schematic representation of two miR-125-centered ceRNET in BC. (A): a simple ceRNET having only one axis, where a pseudogene (CYP4Z2P) mRNA inhibits miR-125a-3p action on the target (CYP4Z1) mRNA by sponging it, thus enhancing CYP4Z1 expression. (B): a more complex ceRNET in which the interaction between miR-125b and STARD13 mRNA controls the expression of multiple target mRNA. See text for references and further explanations.
In 2004, Rieger and collaborators discovered a new human cytochrome P450 (CYP), termed CYP4Z1, which is specifically expressed in mammary gland and breast carcinoma [142]. They also found a transcribed pseudogene, named CYP4Z2P, that codes for a truncated CYP protein (340 amino acids vs. 505) with 96% identity to CYP4Z1. Both CYPs are highly expressed in BC, although the expression level of CYP4Z2P is approximately 20 times lower than that of CYP4Z1 in mammary tissues and barely expressed elsewhere. Later, it was shown that increased expression of CYP4Z1 promotes tumor angiogenesis and growth in human BC [143] and that CYP4Z2P 3′-UTR is involved in promoting BC angiogenesis through the VEGF/VEGFR2 pathway [144]. In 2015, Zheng et al. showed that the action of CYP4Z2P 3′-UTR is sponging several miRs, including miR-125a-3p, and that this pseudogene acts as a ceRNA with respect to CYP4Z1 mRNA, enhancing its expression levels [145]. They also showed that tumor angiogenesis is promoted by overexpression of the CYP4Z2P and CYP4Z1-3′UTRs, which significantly increased the activation of the ERK1/2 and PI3K/Akt pathways through the induction of their phosphorylation. The same group also showed a number of interactions later: (i) deregulation of these ceRNA also confers tamoxifen resistance in BC through the enhancement of the transcriptional activity of ERα via its phosphorylation dependent on cyclin-dependent kinase 3 (CDK3) [146]; (ii) downregulation of CYP4Z1 or CYP4Z2P through 3′-UTR binding promotes cell apoptosis, mirroring the functions and modulating the expression of human telomerase reverse transcriptase (hTERT) [147]; (iii) transcriptional factor six2 activates these CYPs ceRNET by directly binding to their promoters, thus activating the downstream PI3K/Akt and ERK1/2 pathways and consequently being involved not only in chemoresistance but also regulating the stemness of BC cells [148].
STARD13 (StAR-related lipid transfer domain protein 13, also known as deleted in liver cancer 2 protein (DLC-2)) is a Rho GTPase-activating protein (Rho GAP) that selectively activates RhoA and CDC42 and suppresses cell growth by inhibiting actin stress fiber assembly in hepatocellular carcinoma (HCC) [149]; this protein is ubiquitously expressed in normal tissues and downregulated in HCC. In mice, STARD13 promotes angiogenesis through the actions of RhoA [150]. Its role is well established in BC as well, where it acts as a tumor suppressor gene [151], regulates cell motility and invasion [152], endothelial differentiation [45], metastasis formation [153][154], cell migration [155], and apoptosis [156]. It has also been shown that STARD13 exerts its function in BC through its participation in many ceRNETs, such as the one involving a positive TGF-β/miR-9 regulatory loop mediated by the STARD13/YAP axis [157], the one involving hsa-miR-21-3p [158], or even the more complex network that involves five different miRs and that controls YAP/TAZ nuclear accumulation and transcriptional activity via modulation of Hippo and Rho-GTPase/F-actin signaling pathways [159]. A direct link between miR-125 and STARD13 expression has been described, too. Li and coworkers showed that CDH5, HOXD1, and HOXD10 encode putative STARD13 ceRNA and display concordant patterns with STARD13 in different metastatic potential BC cell lines and tissues; in addition, they also show that the 3′ UTR of STARD13 mRNA can bind miR-125b (and also miR-9 and miR-10b), indicating that this mRNA may participate in multiple pathways simultaneously [160], thus confirming their previous study about this interaction [132] and showing that the transcripts of the tumor suppressor genes CDH5, HOXD1, and HOXD10 inhibit BC metastasis in vitro and in vivo by competing with STARD13 mRNA for these three miR. Interestingly, CDH5, HOXD1 and HOXD10, along with STARD13, are BC players also in a different ceRNET, competing for a different set of miRs, indicating that STARD13′s role in BC is very complex. In 2017, Hu et al. discovered another ceRNET axis in which STARD13 and miR-125b control CCR2 (cysteine–cysteine chemokine receptor 2) expression levels [153]. In this case, the authors found that the CCR2 3′ UTR harbors three miR-125 binding sites that both inhibit MDA-MB-231 and MCF-7 cell metastasis by repressing epithelial-mesenchymal transition (EMT) in vitro and suppress BC metastasis in vivo through competition with STARD13 in a miR-125b-dependent and protein-coding-independent manner. Another component of the same ceRNET is TP53INP1 (tumor protein p53-inducible nuclear protein 1). TP53INP1 is an antiproliferative and proapoptotic protein involved in cell stress response that acts as a dual regulator of transcription and autophagy and is modulated by p53 in response to stress; it also interacts with kinases HIPK2 and PKCδ, which phosphorylate p53, creating a positive feedback loop between p53 and TP53INP1 [161]. TP53INP1 is also involved in SPARC (secreted protein acidic and rich in cysteine)-mediated-promotive effects on cancer cell migration and metastasis [162]. In 2018, Zheng et al. found a ceRNA interaction between STARD13 and TP53INP1 mediated by competitively binding to miR-125b in BC [163]. In this case, STARD13 promotes upregulation of TP53INP1, causing the inhibition of BC cell metastasis through competitive binding to miR-125b thanks to the inhibition of SPARC gene expression. Later, Guo and co-workers also found a ceRNET axis in BC involving miR-125b, STARD13 and BMF (Bcl-2-modifying factor) mRNA [156]. BMF is a member of the BCL2 protein family and controls apoptosis in several cell types [164]. The authors [156] found that miR-125b directly binds the 3′ UTR and thus downregulates BMF expression, and that STARD13, sponging miR-125b, upregulates BMF in BC both in vitro and in vivo. All together, these results suggest novel therapies for BC treatment and aid in selecting adequate drugs, depending on the molecular biology of the tumor, from a perspective aiming at the goal of personalized medicine. Indeed, a recent study showed that tanshinone IIA (an effective component extracted from Salvia miltiorrhiza that regulates the stemness of tumor cells) attenuates this phenotype in BC cells by downregulating miR-125b levels and upregulating its target gene STARD13 expression, while miR-125b overexpression or STARD13 knockdown impairs the inhibitory effects of tanshinone IIA on the stemness of BC cells [165].
References
- Naeli, P.; Winter, T.; Hackett, A.P.; Alboushi, L.; Jafarnejad, S.M. The Intricate Balance between MicroRNA-Induced MRNA Decay and Translational Repression. FEBS J. 2023, 290, 2508–2524.
- Friedman, R.C.; Farh, K.K.H.; Burge, C.B.; Bartel, D.P. Most Mammalian MRNAs Are Conserved Targets of MicroRNAs. Genome Res. 2009, 19, 92–105.
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Marshall, M.; et al. A Uniform System for MicroRNA Annotation. RNA 2003, 9, 277–279.
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. MiRBase: MicroRNA Sequences, Targets and Gene Nomenclature. Nucleic Acids Res. 2006, 34, D140–D144.
- Dziechciowska, I.; Dąbrowska, M.; Mizielska, A.; Pyra, N.; Lisiak, N.; Kopczyński, P.; Jankowska-Wajda, M.; Rubiś, B. MiRNA Expression Profiling in Human Breast Cancer Diagnostics and Therapy. Curr. Issues Mol. Biol. 2023, 45, 9500–9525.
- Loh, H.Y.; Norman, B.P.; Lai, K.S.; Rahman, N.M.A.N.A.; Alitheen, N.B.M.; Osman, M.A. The Regulatory Role of MicroRNAs in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 4940.
- Najjary, S.; Mohammadzadeh, R.; Mokhtarzadeh, A.; Mohammadi, A.; Kojabad, A.B.; Baradaran, B. Role of MiR-21 as an Authentic Oncogene in Mediating Drug Resistance in Breast Cancer. Gene 2020, 738, 144453.
- Wang, Z.X.; Lu, B.B.; Wang, H.; Cheng, Z.X.; Yin, Y.M. MicroRNA-21 Modulates Chemosensitivity of Breast Cancer Cells to Doxorubicin by Targeting PTEN. Arch. Med. Res. 2011, 42, 281–290.
- Wang, H.; Tan, Z.; Hu, H.; Liu, H.; Wu, T.; Zheng, C.; Wang, X.; Luo, Z.; Wang, J.; Liu, S.; et al. MicroRNA-21 Promotes Breast Cancer Proliferation and Metastasis by Targeting LZTFL1. BMC Cancer 2019, 19, 738.
- Shi, Y.; Ye, P.; Long, X. Differential Expression Profiles of the Transcriptome in Breast Cancer Cell Lines Revealed by Next Generation Sequencing. Cell. Physiol. Biochem. 2017, 44, 804–816.
- Mohmmed, E.A.; Shousha, W.G.; EL-Saiid, A.S.; Ramadan, S.S. A Clinical Evaluation of Circulating MiR-106a and Raf-1 as Breast Cancer Diagnostic and Prognostic Markers. Asian Pac. J. Cancer Prev. 2021, 22, 3513–3520.
- You, F.; Luan, H.; Sun, D.; Cui, T.; Ding, P.; Tang, H.; Sun, D. MiRNA-106a Promotes Breast Cancer Cell Proliferation, Clonogenicity, Migration, and Invasion Through Inhibiting Apoptosis and Chemosensitivity. DNA Cell Biol. 2019, 38, 198–207.
- You, F.; Li, J.; Zhang, P.; Zhang, H.; Cao, X. MiR106a Promotes the Growth of Transplanted Breast Cancer and Decreases the Sensitivity of Transplanted Tumors to Cisplatin. Cancer Manag. Res. 2020, 12, 233–246.
- Dinami, R.; Ercolani, C.; Petti, E.; Piazza, S.; Ciani, Y.; Sestito, R.; Sacconi, A.; Biagioni, F.; Le Sage, C.; Agami, R.; et al. MiR-155 Drives Telomere Fragility in Human Breast Cancer by Targeting TRF1. Cancer Res. 2014, 74, 4145–4156.
- Roth, C.; Rack, B.; Müller, V.; Janni, W.; Pantel, K.; Schwarzenbach, H. Circulating MicroRNAs as Blood-Based Markers for Patients with Primary and Metastatic Breast Cancer. Breast Cancer Res. 2010, 12, R90.
- Li, P.; Xu, T.; Zhou, X.; Liao, L.; Pang, G.; Luo, W.; Han, L.; Zhang, J.; Luo, X.; Xie, X.; et al. Downregulation of MiRNA-141 in Breast Cancer Cells Is Associated with Cell Migration and Invasion: Involvement of ANP32E Targeting. Cancer Med. 2017, 6, 662–672.
- Xiong, Z.; Ye, L.; Zhenyu, H.; Li, F.; Xiong, Y.; Lin, C.; Wu, X.; Deng, G.; Shi, W.; Song, L.; et al. ANP32E Induces Tumorigenesis of Triple-Negative Breast Cancer Cells by Upregulating E2F1. Mol. Oncol. 2018, 12, 896–912.
- Taha, M.; Mitwally, N.; Soliman, A.S.; Yousef, E. Potential Diagnostic and Prognostic Utility of MiR-141, MiR-181b1, and MiR-23b in Breast Cancer. Int. J. Mol. Sci. 2020, 21, 8589.
- Li, X.X.; Gao, S.Y.; Wang, P.Y.; Zhou, X.; Li, Y.J.; Yu, Y.; Yan, Y.F.; Zhang, H.H.; Lv, C.J.; Zhou, H.H.; et al. Reduced Expression Levels of Let-7c in Human Breast Cancer Patients. Oncol. Lett. 2015, 9, 1207–1212.
- Fu, X.; Mao, X.; Wang, Y.; Ding, X.; Li, Y. Let-7c-5p Inhibits Cell Proliferation and Induces Cell Apoptosis by Targeting ERCC6 in Breast Cancer. Oncol. Rep. 2017, 38, 1851–1856.
- Swellam, M.; Mahmoud, M.S.; Hashim, M.; Hassan, N.M.; Sobeih, M.E.; Nageeb, A.M. Clinical Aspects of Circulating MiRNA-335 in Breast Cancer Patients: A Prospective Study. J. Cell. Biochem. 2019, 120, 8975–8982.
- Heyn, H.; Engelmann, M.; Schreek, S.; Ahrens, P.; Lehmann, U.; Kreipe, H.; Schlegelberger, B.; Beger, C. MicroRNA MiR-335 Is Crucial for the BRCA1 Regulatory Cascade in Breast Cancer Development. Int. J. Cancer 2011, 129, 2797–2806.
- Gao, Y.; Zeng, F.; Wu, J.Y.; Li, H.Y.; Fan, J.J.; Mai, L.; Zhang, J.; Ma, D.M.; Li, Y.; Song, F.Z. MiR-335 Inhibits Migration of Breast Cancer Cells through Targeting Oncoprotein c-Met. Tumour Biol. 2015, 36, 2875–2883.
- Soofiyani, S.R.; Hosseini, K.; Ebrahimi, T.; Forouhandeh, H.; Sadeghi, M.; Beirami, S.M.; Ghasemnejad, T.; Tarhriz, V.; Montazersaheb, S. Prognostic Value and Biological Role of MiR-126 in Breast Cancer. MicroRNA 2022, 11, 95–103.
- Zhu, N.; Zhang, D.; Xie, H.; Zhou, Z.; Chen, H.; Hu, T.; Bai, Y.; Shen, Y.; Yuan, W.; Jing, Q.; et al. Endothelial-Specific Intron-Derived MiR-126 Is down-Regulated in Human Breast Cancer and Targets Both VEGFA and PIK3R2. Mol. Cell. Biochem. 2011, 351, 157–164.
- Fu, R.; Tong, J.S. MiR-126 Reduces Trastuzumab Resistance by Targeting PIK3R2 and Regulating AKT/MTOR Pathway in Breast Cancer Cells. J. Cell. Mol. Med. 2020, 24, 7600–7608.
- Wang, C.Z.; Yuan, P.; Li, Y. MiR-126 Regulated Breast Cancer Cell Invasion by Targeting ADAM9. Int. J. Clin. Exp. Pathol. 2015, 8, 6547–6553.
- Li, S.Q.; Wang, Z.H.; Mi, X.G.; Liu, L.; Tan, Y. MiR-199a/b-3p Suppresses Migration and Invasion of Breast Cancer Cells by Downregulating PAK4/MEK/ERK Signaling Pathway. IUBMB Life 2015, 67, 768–777.
- Qattan, A.; Al-Tweigeri, T.; Alkhayal, W.; Suleman, K.; Tulbah, A.; Amer, S. Clinical Identification of Dysregulated Circulating MicroRNAs and Their Implication in Drug Response in Triple Negative Breast Cancer (TNBC) by Target Gene Network and Meta-Analysis. Genes 2021, 12, 549.
- Fan, X.; Zhou, S.; Zheng, M.; Deng, X.; Yi, Y.; Huang, T. MiR-199a-3p Enhances Breast Cancer Cell Sensitivity to Cisplatin by Downregulating TFAM (TFAM). Biomed. Pharmacother. 2017, 88, 507–514.
- Zuo, Y.; Qu, C.; Tian, Y.; Wen, Y.; Xia, S.; Ma, M. The HIF-1/SNHG1/MiR-199a-3p/TFAM Axis Explains Tumor Angiogenesis and Metastasis under Hypoxic Conditions in Breast Cancer. Biofactors 2021, 47, 444–460.
- Fuso, P.; Di Salvatore, M.; Santonocito, C.; Guarino, D.; Autilio, C.; Mulè, A.; Arciuolo, D.; Rinninella, A.; Mignone, F.; Ramundo, M.; et al. Let-7a-5p, MiR-100-5p, MiR-101-3p, and MiR-199a-3p Hyperexpression as Potential Predictive Biomarkers in Early Breast Cancer Patients. J. Pers. Med. 2021, 11, 816.
- Wang, C.Z.; Deng, F.; Li, H.; Wang, D.D.; Zhang, W.; Ding, L.; Tang, J.H. MiR-101: A Potential Therapeutic Target of Cancers. Am. J. Transl. Res. 2018, 10, 3310–3321.
- Harati, R.; Mohammad, M.G.; Tlili, A.; El-Awady, R.A.; Hamoudi, R. Loss of MiR-101-3p Promotes Transmigration of Metastatic Breast Cancer Cells through the Brain Endothelium by Inducing COX-2/MMP1 Signaling. Pharmaceuticals 2020, 13, 144.
- Jiang, H.; Li, L.; Zhang, J.; Wan, Z.; Wang, Y.; Hou, J.; Yu, Y. MiR-101-3p and Syn-Cal14.1a Synergy in Suppressing EZH2-Induced Progression of Breast Cancer. Onco Targets Ther. 2020, 13, 9599–9609.
- Toda, H.; Seki, N.; Kurozumi, S.; Shinden, Y.; Yamada, Y.; Nohata, N.; Moriya, S.; Idichi, T.; Maemura, K.; Fujii, T.; et al. RNA-sequence-based MicroRNA Expression Signature in Breast Cancer: Tumor-suppressive MiR-101-5p Regulates Molecular Pathogenesis. Mol. Oncol. 2020, 14, 426–446.
- Piasecka, D.; Braun, M.; Kordek, R.; Sadej, R.; Romanska, H. MicroRNAs in Regulation of Triple-Negative Breast Cancer Progression. J. Cancer Res. Clin. Oncol. 2018, 144, 1401–1411.
- Sporn, J.C.; Katsuta, E.; Yan, L.; Takabe, K. Expression of MicroRNA-9 Is Associated with Overall Survival in Breast Cancer Patients. J. Surg. Res. 2019, 233, 426–435.
- Gwak, J.M.; Kim, H.J.; Kim, E.J.; Chung, Y.R.; Yun, S.; Seo, A.N.; Lee, H.J.; Park, S.Y. MicroRNA-9 Is Associated with Epithelial-Mesenchymal Transition, Breast Cancer Stem Cell Phenotype, and Tumor Progression in Breast Cancer. Breast Cancer Res. Treat. 2014, 147, 39–49.
- Shen, M.; Dong, C.; Ruan, X.; Yan, W.; Cao, M.; Pizzo, D.; Wu, X.; Yang, L.; Liu, L.; Ren, X.; et al. Chemotherapy-Induced Extracellular Vesicle miRNAs Promote Breast Cancer Stemness by Targeting ONECUT2. Cancer Res. 2019, 79, 3608–3621.
- Liu, D.Z.; Chang, B.; Li, X.D.; Zhang, Q.H.; Zou, Y.H. MicroRNA-9 Promotes the Proliferation, Migration, and Invasion of Breast Cancer Cells via down-Regulating FOXO1. Clin. Transl. Oncol. 2017, 19, 1133–1140.
- Li, X.; Zeng, Z.; Wang, J.; Wu, Y.; Chen, W.; Zheng, L.; Xi, T.; Wang, A.; Lu, Y. MicroRNA-9 and Breast Cancer. Biomed. Pharmacother. 2020, 122, 109687.
- Chen, D.; Sun, Y.; Wei, Y.; Zhang, P.; Rezaeian, A.H.; Teruya-Feldstein, J.; Gupta, S.; Liang, H.; Lin, H.K.; Hung, M.C.; et al. LIFR Is a Breast Cancer Metastasis Suppressor Upstream of the Hippo-YAP Pathway and a Prognostic Marker. Nat. Med. 2012, 18, 1511–1517.
- Wang, S.; Cheng, M.; Zheng, X.; Zheng, L.; Liu, H.; Lu, J.; Liu, Y.; Chen, W. Interactions Between LncRNA TUG1 and MiR-9-5p Modulate the Resistance of Breast Cancer Cells to Doxorubicin by Regulating EIF5A2. OncoTargets Ther. 2020, 13, 13159–13170.
- D’Ippolito, E.; Plantamura, I.; Bongiovanni, L.; Casalini, P.; Baroni, S.; Piovan, C.; Orlandi, R.; Gualeni, A.V.; Gloghini, A.; Rossini, A.; et al. MiR-9 and MiR-200 Regulate PDGFRβ-Mediated Endothelial Differentiation of Tumor Cells in Triple-Negative Breast Cancer. Cancer Res. 2016, 76, 5562–5572.
- Khew-Goodall, Y.; Goodall, G.J. Myc-Modulated MiR-9 Makes More Metastases. Nat. Cell Biol. 2010, 12, 209–211.
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Onder, T.T.; Valastyan, S.; et al. MiR-9, a MYC/MYCN-Activated MicroRNA, Regulates E-Cadherin and Cancer Metastasis. Nat. Cell Biol. 2010, 12, 247–256.
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 1993, 75, 843–854.
- Duan, R.; Pak, C.H.; Jin, P. Single Nucleotide Polymorphism Associated with Mature MiR-125a Alters the Processing of Pri-MiRNA. Hum. Mol. Genet. 2007, 16, 1124–1131.
- Shaham, L.; Binder, V.; Gefen, N.; Borkhardt, A.; Izraeli, S. MiR-125 in Normal and Malignant Hematopoiesis. Leukemia 2012, 26, 2011–2018.
- Ciafrè, S.A.; Galardi, S.; Mangiola, A.; Ferracin, M.; Liu, C.G.; Sabatino, G.; Negrini, M.; Maira, G.; Croce, C.M.; Farace, M.G. Extensive Modulation of a Set of MicroRNAs in Primary Glioblastoma. Biochem. Biophys. Res. Commun. 2005, 334, 1351–1358.
- Emmrich, S.; Streltsov, A.; Schmidt, F.; Thangapandi, V.R.; Reinhardt, D.; Klusmann, J.H. LincRNAs MONC and MIR100HG Act as Oncogenes in Acute Megakaryoblastic Leukemia. Mol. Cancer 2014, 13, 171.
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of Tissue-Specific MicroRNAs from Mouse. Curr. Biol. 2002, 12, 735–739.
- Huang, H.Y.; Lin, Y.C.D.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. MiRTarBase Update 2022: An Informative Resource for Experimentally Validated MiRNA-Target Interactions. Nucleic Acids Res. 2022, 50, D222–D230.
- MiRTarBase: The Experimentally Validated MicroRNA-Target Interactions Database. Available online: https://mirtarbase.cuhk.edu.cn/~miRTarBase/miRTarBase_2022/php/index.php (accessed on 24 August 2023).
- Sun, Y.M.; Lin, K.Y.; Chen, Y.Q. Diverse Functions of MiR-125 Family in Different Cell Contexts. J. Hematol. Oncol. 2013, 6, 6.
- Ji, X.; Lu, Y.; Tian, H.; Meng, X.; Wei, M.; Cho, W.C. Chemoresistance Mechanisms of Breast Cancer and Their Countermeasures. Biomed. Pharmacother. 2019, 114, 108800.
- Ge, Y.; Sun, Y.; Chen, J. IGF-II Is Regulated by MicroRNA-125b in Skeletal Myogenesis. J. Cell Biol. 2011, 192, 69–81.
- Qiu, J.; Zhu, J.; Zhang, R.; Liang, W.; Ma, W.; Zhang, Q.; Huang, Z.; Ding, F.; Sun, H. MiR-125b-5p Targeting TRAF6 Relieves Skeletal Muscle Atrophy Induced by Fasting or Denervation. Ann. Transl. Med. 2019, 7, 456.
- Wang, X.; Chen, S.; Gao, Y.; Yu, C.; Nie, Z.; Lu, R.; Sun, Y.; Guan, Z. MicroRNA-125b Inhibits the Proliferation of Vascular Smooth Muscle Cells Induced by Platelet-Derived Growth Factor BB. Exp. Ther. Med. 2021, 22, 791.
- Wang, Y.; Tan, J.; Wang, L.; Pei, G.; Cheng, H.; Zhang, Q.; Wang, S.; He, C.; Fu, C.; Wei, Q. MiR-125 Family in Cardiovascular and Cerebrovascular Diseases. Front. Cell Dev. Biol. 2021, 9, 799049.
- Lozano-Velasco, E.; Galiano-Torres, J.; Jodar-Garcia, A.; Aranega, A.E.; Franco, D. MiR-27 and MiR-125 Distinctly Regulate Muscle-Enriched Transcription Factors in Cardiac and Skeletal Myocytes. BioMed Res. Int. 2015, 2015, 391306.
- Li, L.; Wang, Q.; Yuan, Z.; Chen, A.; Liu, Z.; Wang, Z.; Li, H. LncRNA-MALAT1 Promotes CPC Proliferation and Migration in Hypoxia by up-Regulation of JMJD6 via Sponging MiR-125. Biochem. Biophys. Res. Commun. 2018, 499, 711–718.
- Li, L.; Zhang, M.; Chen, W.; Wang, R.; Ye, Z.; Wang, Y.; Li, X.; Cai, C. LncRNA-HOTAIR Inhibition Aggravates Oxidative Stress-Induced H9c2 Cells Injury through Suppression of MMP2 by MiR-125. Acta Biochim. Biophys. Sin. 2018, 50, 996–1006.
- Chen, C.Y.; Lee, D.S.; Choong, O.K.; Chang, S.K.; Hsu, T.; Nicholson, M.W.; Liu, L.W.; Lin, P.J.; Ruan, S.C.; Lin, S.W.; et al. Cardiac-Specific MicroRNA-125b Deficiency Induces Perinatal Death and Cardiac Hypertrophy. Sci. Rep. 2021, 11, 2377.
- Le, M.T.N.; Xie, H.; Zhou, B.; Chia, P.H.; Rizk, P.; Um, M.; Udolph, G.; Yang, H.; Lim, B.; Lodish, H.F. MicroRNA-125b Promotes Neuronal Differentiation in Human Cells by Repressing Multiple Targets. Mol. Cell. Biol. 2009, 29, 5290–5305.
- Dash, S.; Balasubramaniam, M.; Rana, T.; Godino, A.; Peck, E.G.; Goodwin, J.S.; Villalta, F.; Calipari, E.S.; Nestler, E.J.; Dash, C.; et al. Poly (ADP-Ribose) Polymerase-1 (PARP-1) Induction by Cocaine Is Post-Transcriptionally Regulated by MiR-125b. eNeuro 2017, 4, ENEURO.0089-17.2017.
- Edbauer, D.; Neilson, J.R.; Foster, K.A.; Wang, C.F.; Seeburg, D.P.; Batterton, M.N.; Tada, T.; Dolan, B.M.; Sharp, P.A.; Sheng, M. Regulation of Synaptic Structure and Function by FMRP-Associated MicroRNAs MiR-125b and MiR-132. Neuron 2010, 65, 373–384.
- Åkerblom, M.; Petri, R.; Sachdeva, R.; Klussendorf, T.; Mattsson, B.; Gentner, B.; Jakobsson, J. MicroRNA-125 Distinguishes Developmentally Generated and Adult-Born Olfactory Bulb Interneurons. Development 2014, 141, 1580–1588.
- Gioia, U.; Di Carlo, V.; Caramanica, P.; Toselli, C.; Cinquino, A.; Marchioni, M.; Laneve, P.; Biagioni, S.; Bozzoni, I.; Cacci, E.; et al. Mir-23a and Mir-125b Regulate Neural Stem/Progenitor Cell Proliferation by Targeting Musashi1. RNA Biol. 2014, 11, 1105–1112.
- Pogue, A.I.; Cui, J.G.; Li, Y.Y.; Zhao, Y.; Culicchia, F.; Lukiw, W.J. Micro RNA-125b (MiRNA-125b) Function in Astrogliosis and Glial Cell Proliferation. Neurosci. Lett. 2010, 476, 18–22.
- Yuan, M.; Da Silva, A.C.A.L.; Arnold, A.; Okeke, L.; Ames, H.; Correa-Cerro, L.S.; Vizcaino, M.A.; Ho, C.Y.; Eberhart, C.G.; Rodriguez, F.J. MicroRNA (MiR) 125b Regulates Cell Growth and Invasion in Pediatric Low Grade Glioma. Sci. Rep. 2018, 8, 12506.
- Laneve, P.; Di Marcotullio, L.; Gioia, U.; Fiori, M.E.; Ferretti, E.; Gulino, A.; Bozzoni, I.; Caffarelli, E. The Interplay between MicroRNAs and the Neurotrophin Receptor Tropomyosin-Related Kinase C Controls Proliferation of Human Neuroblastoma Cells. Proc. Natl. Acad. Sci. USA 2007, 104, 7957–7962.
- Ferretti, E.; De Smaele, E.; Po, A.; Marcotullio, L.D.; Tosi, E.; Espinola, M.S.B.; Rocco, C.D.; Riccardi, R.; Giangaspero, F.; Farcomeni, A.; et al. MicroRNA Profiling in Human Medulloblastoma. Int. J. Cancer 2009, 124, 568–577.
- Wu, N.; Lin, X.; Zhao, X.; Zheng, L.; Xiao, L.; Liu, J.; Ge, L.; Cao, S. MiR-125b Acts as an Oncogene in Glioblastoma Cells and Inhibits Cell Apoptosis through P53 and P38MAPK-Independent Pathways. Br. J. Cancer 2013, 109, 2853–2863.
- Xia, H.F.; He, T.Z.; Liu, C.M.; Cui, Y.; Song, P.P.; Jin, X.H.; Ma, X. MiR-125b Expression Affects the Proliferation and Apoptosis of Human Glioma Cells by Targeting Bmf. Cell. Physiol. Biochem. 2009, 23, 347–358.
- Wojtowicz, E.E.; Lechman, E.R.; Hermans, K.G.; Schoof, E.M.; Wienholds, E.; Isserlin, R.; van Veelen, P.A.; Broekhuis, M.J.C.; Janssen, G.M.C.; Trotman-Grant, A.; et al. Ectopic MiR-125a Expression Induces Long-Term Repopulating Stem Cell Capacity in Mouse and Human Hematopoietic Progenitors. Cell Stem Cell 2016, 19, 383–396.
- Guo, S.; Lu, J.; Schlanger, R.; Zhang, H.; Wang, J.Y.; Fox, M.C.; Purton, L.E.; Fleming, H.H.; Cobb, B.; Merkenschlager, M.; et al. MicroRNA MiR-125a Controls Hematopoietic Stem Cell Number. Proc. Natl. Acad. Sci. USA 2010, 107, 14229–14234.
- Emmrich, S.; Rasche, M.; Schöning, J.; Reimer, C.; Keihani, S.; Maroz, A.; Xie, Y.; Li, Z.; Schambach, A.; Reinhardt, D.; et al. MiR-99a/100~125b Tricistrons Regulate Hematopoietic Stem and Progenitor Cell Homeostasis by Shifting the Balance between TGFβ and Wnt Signaling. Genes Dev. 2014, 28, 858–874.
- Allantaz, F.; Cheng, D.T.; Bergauer, T.; Ravindran, P.; Rossier, M.F.; Ebeling, M.; Badi, L.; Reis, B.; Bitter, H.; D’Asaro, M.; et al. Expression Profiling of Human Immune Cell Subsets Identifies MiRNA-MRNA Regulatory Relationships Correlated with Cell Type Specific Expression. PLoS ONE 2012, 7, e29979.
- Yao, D.; Zhou, Z.; Wang, P.; Zheng, L.; Huang, Y.; Duan, Y.; Liu, B.; Li, Y. MiR-125-5p/IL-6R Axis Regulates Macrophage Inflammatory Response and Intestinal Epithelial Cell Apoptosis in Ulcerative Colitis through JAK1/STAT3 and NF-ΚB Pathway. Cell Cycle 2021, 20, 2547–2564.
- Yu, C.; Tang, W.; Lu, R.; Tao, Y.; Ren, T.; Gao, Y. Human Adipose-Derived Mesenchymal Stem Cells Promote Lymphocyte Apoptosis and Alleviate Atherosclerosis via MiR-125b-1-3p/BCL11B Signal Axis. Ann. Palliat. Med. 2021, 10, 2123–2133.
- Sun, X.; Zhang, S.; Ma, X. Prognostic Value of MicroRNA-125 in Various Human Malignant Neoplasms: A Meta-Analysis. Clin. Lab. 2015, 61, 1667–1674.
- Cowden Dahl, K.D.; Dahl, R.; Kruichak, J.N.; Hudson, L.G. The Epidermal Growth Factor Receptor Responsive MiR-125a Represses Mesenchymal Morphology in Ovarian Cancer Cells. Neoplasia 2009, 11, 1208–1215.
- Guan, Y.; Yao, H.; Zheng, Z.; Qiu, G.; Sun, K. MiR-125b Targets BCL3 and Suppresses Ovarian Cancer Proliferation. Int. J. Cancer 2011, 128, 2274–2283.
- Chen, Z.; Guo, X.; Sun, S.; Lu, C.; Wang, L. Serum MiR-125b Levels Associated with Epithelial Ovarian Cancer (EOC) Development and Treatment Responses. Bioengineered 2020, 11, 311–317.
- Huang, L.; Luo, J.; Cai, Q.; Pan, Q.; Zeng, H.; Guo, Z.; Dong, W.; Huang, J.; Lin, T. MicroRNA-125b Suppresses the Development of Bladder Cancer by Targeting E2F3. Int. J. Cancer 2011, 128, 1758–1769.
- Pospisilova, S.; Pazzourkova, E.; Horinek, A.; Brisuda, A.; Svobodova, I.; Soukup, V.; Hrbacek, J.; Capoun, O.; Hanus, T.; Mares, J.; et al. MicroRNAs in Urine Supernatant as Potential Non-Invasive Markers for Bladder Cancer Detection. Neoplasma 2016, 63, 799–808.
- Blick, C.; Ramachandran, A.; Mccormick, R.; Wigfield, S.; Cranston, D.; Catto, J.; Harris, A.L. Identification of a Hypoxia-Regulated MiRNA Signature in Bladder Cancer and a Role for MiR-145 in Hypoxia-Dependent Apoptosis. Br. J. Cancer 2015, 113, 634–644.
- Zhou, H.; Tang, K.; Xiao, H.; Zeng, J.; Guan, W.; Guo, X.; Xu, H.; Ye, Z. A Panel of Eight-MiRNA Signature as a Potential Biomarker for Predicting Survival in Bladder Cancer. J. Exp. Clin. Cancer Res. 2015, 34, 53.
- Bi, Q.; Tang, S.; Xia, L.; Du, R.; Fan, R.; Gao, L.; Jin, J.; Liang, S.; Chen, Z.; Xu, G.; et al. Ectopic Expression of MiR-125a Inhibits the Proliferation and Metastasis of Hepatocellular Carcinoma by Targeting MMP11 and VEGF. PLoS ONE 2012, 7, e40169.
- Jia, H.Y.; Wang, Y.X.; Yan, W.T.; Li, H.Y.; Tian, Y.Z.; Wang, S.M.; Zhao, H.L. MicroRNA-125b Functions as a Tumor Suppressor in Hepatocellular Carcinoma Cells. Int. J. Mol. Sci. 2012, 13, 8762–8774.
- Liang, L.; Wong, C.M.; Ying, Q.; Fan, D.N.Y.; Huang, S.; Ding, J.; Yao, J.; Yan, M.; Li, J.; Yao, M.; et al. MicroRNA-125b Suppressesed Human Liver Cancer Cell Proliferation and Metastasis by Directly Targeting Oncogene LIN28B2. Hepatology 2010, 52, 1731–1740.
- Kong, J.; Liu, X.; Li, X.; Wu, J.; Wu, N.; Chen, J.; Fang, F. MiR-125/Pokemon Auto-Circuit Contributes to the Progression of Hepatocellular Carcinoma. Tumour Biol. 2016, 37, 511–519.
- Xie, C.; Zhang, L.Z.; Chen, Z.L.; Zhong, W.J.; Fang, J.H.; Zhu, Y.; Xiao, M.H.; Guo, Z.W.; Zhao, N.; He, X.; et al. A HMTR4-PDIA3P1-MiR-125/124-TRAF6 Regulatory Axis and Its Function in NF Kappa B Signaling and Chemoresistance. Hepatology 2020, 71, 1660–1677.
- Jiang, J.X.; Gao, S.; Pan, Y.Z.; Yu, C.; Sun, C.Y. Overexpression of MicroRNA-125b Sensitizes Human Hepatocellular Carcinoma Cells to 5-Fluorouracil through Inhibition of Glycolysis by Targeting Hexokinase II. Mol. Med. Rep. 2014, 10, 995–1002.
- Xu, Z.; Pei, C.; Cheng, H.; Song, K.; Yang, J.; Li, Y.; He, Y.; Liang, W.; Liu, B.; Tan, W.; et al. Comprehensive Analysis of FOXM1 Immune Infiltrates, M6a, Glycolysis and CeRNA Network in Human Hepatocellular Carcinoma. Front. Immunol. 2023, 14, 1138524.
- Chen, W.; Wang, T.; Li, W.; Yin, S. MiR-125b Acts as a Tumor Suppressor of Melanoma by Targeting NCAM. JBUON 2021, 26, 182–188.
- Kappelmann, M.; Kuphal, S.; Meister, G.; Vardimon, L.; Bosserhoff, A.K. MicroRNA MiR-125b Controls Melanoma Progression by Direct Regulation of c-Jun Protein Expression. Oncogene 2013, 32, 2984–2991.
- Xu, N.; Zhang, L.; Meisgen, F.; Harada, M.; Heilborn, J.; Homey, B.; Grandér, D.; Ståhle, M.; Sonkoly, E.; Pivarcsi, A. MicroRNA-125b down-Regulates Matrix Metallopeptidase 13 and Inhibits Cutaneous Squamous Cell Carcinoma Cell Proliferation, Migration, and Invasion. J. Biol. Chem. 2012, 287, 29899–29908.
- Tian, K.; Liu, W.; Zhang, J.; Fan, X.; Liu, J.; Zhao, N.; Yao, C.; Miao, G. MicroRNA-125b Exerts Antitumor Functions in Cutaneous Squamous Cell Carcinoma by Targeting the STAT3 Pathway. Cell. Mol. Biol. Lett. 2020, 25, 12.
- Sand, M.; Skrygan, M.; Sand, D.; Georgas, D.; Hahn, S.A.; Gambichler, T.; Altmeyer, P.; Bechara, F.G. Expression of MicroRNAs in Basal Cell Carcinoma. Br. J. Dermatol. 2012, 167, 847–855.
- Liu, L.H.; Li, H.; Li, J.P.; Zhong, H.; Zhang, H.C.; Chen, J.; Xiao, T. MiR-125b Suppresses the Proliferation and Migration of Osteosarcoma Cells through down-Regulation of STAT3. Biochem. Biophys. Res. Commun. 2011, 416, 31–38.
- Yang, Y.; Chen, Y.; Liu, J.; Zhang, B.; Yang, L.; Xue, J.; Zhang, Z.; Qin, L.; Bian, R. MiR-125b-5p/STAT3 Axis Regulates Drug Resistance in Osteosarcoma Cells by Acting on ABC Transporters. Stem Cells Int. 2023, 2023, 9997676.
- Tang, X.Y.; Zheng, W.; Ding, M.; Guo, K.J.; Yuan, F.; Feng, H.; Deng, B.; Sun, W.; Hou, Y.; Gao, L. MiR-125b Acts as a Tumor Suppressor in Chondrosarcoma Cells by the Sensitization to Doxorubicin through Direct Targeting the ErbB2-Regulated Glucose Metabolism. Drug Des. Dev. Ther. 2016, 10, 571–583.
- Gao, S.; Sun, H.; Cheng, C.; Wang, G. BRCA1-Associated Protein-1 Suppresses Osteosarcoma Cell Proliferation and Migration Through Regulation PI3K/Akt Pathway. DNA Cell Biol. 2017, 36, 386–393.
- Wu, S.; Shen, W.; Yang, L.; Zhu, M.; Zhang, M.; Zong, F.; Geng, L.; Wang, Y.; Huang, T.; Pan, Y.; et al. Genetic Variations in MiR-125 Family and the Survival of Non-Small Cell Lung Cancer in Chinese Population. Cancer Med. 2019, 8, 2636–2645.
- Wang, G.; Mao, W.; Zheng, S.; Ye, J. Epidermal Growth Factor Receptor-Regulated MiR-125a-5p--a Metastatic Inhibitor of Lung Cancer. FEBS J. 2009, 276, 5571–5578.
- Yagishita, S.; Fujita, Y.; Kitazono, S.; Ko, R.; Nakadate, Y.; Sawada, T.; Kitamura, Y.; Shimoyama, T.; Maeda, Y.; Takahashi, F.; et al. Chemotherapy-Regulated MicroRNA-125-HER2 Pathway as a Novel Therapeutic Target for Trastuzumab-Mediated Cellular Cytotoxicity in Small Cell Lung Cancer. Mol. Cancer Ther. 2015, 14, 1414–1423.
- Yu, X.; Wei, F.; Yu, J.; Zhao, H.; Jia, L.; Ye, Y.; Du, R.; Ren, X.; Li, H. Matrix Metalloproteinase 13: A Potential Intermediate between Low Expression of MicroRNA-125b and Increasing Metastatic Potential of Non-Small Cell Lung Cancer. Cancer Genet. 2015, 208, 76–84.
- Bloomston, M.; Frankel, W.L.; Petrocca, F.; Volinia, S.; Alder, H.; Hagan, J.P.; Liu, C.G.; Bhatt, D.; Taccioli, C.; Croce, C.M. MicroRNA Expression Patterns to Differentiate Pancreatic Adenocarcinoma from Normal Pancreas and Chronic Pancreatitis. JAMA 2007, 297, 1901–1908.
- Wang, J.; Paris, P.L.; Chen, J.; Ngo, V.; Yao, H.; Frazier, M.L.; Killary, A.M.; Liu, C.G.; Liang, H.; Mathy, C.; et al. Next Generation Sequencing of Pancreatic Cyst Fluid MicroRNAs from Low Grade-Benign and High Grade-Invasive Lesions. Cancer Lett. 2015, 356, 404–409.
- Xue, Y.; Zhong, Y.; Wu, T.; Sheng, Y.; Dai, Y.; Xu, L.; Bao, C. Anti-Proliferative and Apoptosis-Promoting Effect of MicroRNA-125b on Pancreatic Cancer by Targeting NEDD9 via PI3K/AKT Signaling. Cancer Manag. Res. 2020, 12, 7363–7373.
- Walter, B.A.; Valera, V.A.; Pinto, P.A.; Merino, M.J. Comprehensive MicroRNA Profiling of Prostate Cancer. J. Cancer 2013, 4, 350–357.
- Li, W.; Dong, Y.; Wang, K.J.; Deng, Z.; Zhang, W.; Shen, H.F. Plasma Exosomal MiR-125a-5p and MiR-141-5p as Non-Invasive Biomarkers for Prostate Cancer. Neoplasma 2020, 67, 1314–1318.
- Konoshenko, M.Y.; Lekchnov, E.A.; Bryzgunova, O.E.; Zaporozhchenko, I.A.; Yarmoschuk, S.V.; Pashkovskaya, O.A.; Pak, S.V.; Laktionov, P.P. The Panel of 12 Cell-Free MicroRNAs as Potential Biomarkers in Prostate Neoplasms. Diagnostics 2020, 10, 38.
- Shi, X.B.; Xue, L.; Yang, J.; Ma, A.H.; Zhao, J.; Xu, M.; Tepper, C.G.; Evans, C.P.; Kung, H.J.; White, R.W.D.V. An Androgen-Regulated MiRNA Suppresses Bak1 Expression and Induces Androgen-Independent Growth of Prostate Cancer Cells. Proc. Natl. Acad. Sci. USA 2007, 104, 19983–19988.
- Shi, X.B.; Xue, L.; Ma, A.H.; Tepper, C.G.; Kung, H.J.; White, R.W.D. MiR-125b Promotes Growth of Prostate Cancer Xenograft Tumor through Targeting pro-Apoptotic Genes. Prostate 2011, 71, 538–549.
- Wang, S.; Wu, J.; Ren, J.; Vlantis, A.C.; Li, M.-y.; Liu, S.Y.W.; Ng, E.K.W.; Chan, A.B.W.; Luo, D.C.; Liu, Z.; et al. MicroRNA-125b Interacts with Foxp3 to Induce Autophagy in Thyroid Cancer. Mol. Ther. 2018, 26, 2295–2303.
- Wu, J.G.; Wang, J.J.; Jiang, X.; Lan, J.P.; He, X.J.; Wang, H.J.; Ma, Y.Y.; Xia, Y.J.; Ru, G.Q.; Ma, J.; et al. MiR-125b Promotes Cell Migration and Invasion by Targeting PPP1CA-Rb Signal Pathways in Gastric Cancer, Resulting in a Poor Prognosis. Gastric Cancer 2015, 18, 729–739.
- Tong, Z.; Liu, N.; Lin, L.; Guo, X.; Yang, D.; Zhang, Q. MiR-125a-5p Inhibits Cell Proliferation and Induces Apoptosis in Colon Cancer via Targeting BCL2, BCL2L12 and MCL1. Biomed. Pharmacother. 2015, 75, 129–136.
- Fu, Q.; Liu, Z.; Pan, D.; Zhang, W.; Xu, L.; Zhu, Y.; Liu, H.; Xu, J. Tumor MiR-125b Predicts Recurrence and Survival of Patients with Clear-Cell Renal Cell Carcinoma after Surgical Resection. Cancer Sci. 2014, 105, 1427–1434.
- Mattie, M.D.; Benz, C.C.; Bowers, J.; Sensinger, K.; Wong, L.; Scott, G.K.; Fedele, V.; Ginzinger, D.; Getts, R.; Haqq, C. Optimized High-Throughput MicroRNA Expression Profiling Provides Novel Biomarker Assessment of Clinical Prostate and Breast Cancer Biopsies. Mol. Cancer 2006, 5, 24.
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA Gene Expression Deregulation in Human Breast Cancer. Cancer Res. 2005, 65, 7065–7070.
- Mar-Aguilar, F.; Luna-Aguirre, C.M.; Moreno-Rocha, J.C.; Araiza-Chávez, J.; Trevino, V.; Rodríguez-Padilla, C.; Reséndez-Pérez, D. Differential Expression of MiR-21, MiR-125b and MiR-191 in Breast Cancer Tissue. Asia Pac. J. Clin. Oncol. 2013, 9, 53–59.
- Liang, F.; Yang, M.; Tong, N.; Fang, J.; Pan, Y.; Li, J.; Zhang, X. Identification of Six Key MiRNAs Associated with Breast Cancer through Screening Large-Scale Microarray Data. Oncol. Lett. 2018, 16, 4159–4168.
- Braicu, C.; Raduly, L.; Morar-Bolba, G.; Cojocneanu, R.; Jurj, A.; Pop, L.A.; Pileczki, V.; Ciocan, C.; Moldovan, A.; Irimie, A.; et al. Aberrant MiRNAs Expressed in HER-2 Negative Breast Cancers Patient. J. Exp. Clin. Cancer Res. 2018, 37, 257.
- Incoronato, M.; Grimaldi, A.M.; Mirabelli, P.; Cavaliere, C.; Parente, C.A.; Franzese, M.; Staibano, S.; Ilardi, G.; Russo, D.; Soricelli, A.; et al. Circulating MiRNAs in Untreated Breast Cancer: An Exploratory Multimodality Morpho-Functional Study. Cancers 2019, 11, 876.
- Scott, G.K.; Goga, A.; Bhaumik, D.; Berger, C.E.; Sullivan, C.S.; Benz, C.C. Coordinate Suppression of ERBB2 and ERBB3 by Enforced Expression of Micro-RNA MiR-125a or MiR-125b. J. Biol. Chem. 2007, 282, 1479–1486.
- Zhang, Y.; Yan, L.X.; Wu, Q.N.; Du, Z.M.; Chen, J.; Liao, D.Z.; Huang, M.Y.; Hou, J.H.; Wu, Q.L.; Zeng, M.S.; et al. MiR-125b Is Methylated and Functions as a Tumor Suppressor by Regulating the ETS1 Proto-Oncogene in Human Invasive Breast Cancer. Cancer Res. 2011, 71, 3552–3562.
- Rajabi, H.; Jin, C.; Ahmad, R.; McClary, A.C.; Joshi, M.D.; Kufe, D. Mucin 1 Oncoprotein Expression Is Suppressed by the miR-125b Oncomir. Genes Cancer 2010, 1, 62–68.
- Tang, F.; Zhang, R.; He, Y.; Zou, M.; Guo, L.; Xi, T. MicroRNA-125b Induces Metastasis by Targeting STARD13 in MCF-7 and MDA-MB-231 Breast Cancer Cells. PLoS ONE 2012, 7, e35435.
- Metheetrairut, C.; Adams, B.D.; Nallur, S.; Weidhaas, J.B.; Slack, F.J. Cel-Mir-237 and Its Homologue, Hsa-MiR-125b, Modulate the Cellular Response to Ionizing Radiation. Oncogene 2017, 36, 512–524.
- Wang, H.; Tan, G.; Dong, L.; Cheng, L.; Li, K.; Wang, Z.; Luo, H. Circulating MiR-125b as a Marker Predicting Chemoresistance in Breast Cancer. PLoS ONE 2012, 7, e34210.
- Zhou, M.; Liu, Z.; Zhao, Y.; Ding, Y.; Liu, H.; Xi, Y.; Xiong, W.; Li, G.; Lu, J.; Fodstad, O.; et al. MicroRNA-125b Confers the Resistance of Breast Cancer Cells to Paclitaxel through Suppression of pro-Apoptotic Bcl-2 Antagonist Killer 1 (Bak1) Expression. J. Biol. Chem. 2010, 285, 21496–21507.
- He, H.; Xu, F.; Huang, W.; Luo, S.Y.; Lin, Y.T.; Zhang, G.H.; Du, Q.; Duan, R.H. MiR-125a-5p Expression Is Associated with the Age of Breast Cancer Patients. Genet. Mol. Res. 2015, 14, 17927–17933.
- Wang, Y.; Wei, Y.; Fan, X.; Zhang, P.; Wang, P.; Cheng, S.; Zhang, J. MicroRNA-125b as a Tumor Suppressor by Targeting MMP11 in Breast Cancer. Thorac. Cancer 2020, 11, 1613–1620.
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 388354.
- Abdollahzadeh, R.; Daraei, A.; Mansoori, Y.; Sepahvand, M.; Amoli, M.M.; Tavakkoly-Bazzaz, J. Competing Endogenous RNA (CeRNA) Cross Talk and Language in CeRNA Regulatory Networks: A New Look at Hallmarks of Breast Cancer. J. Cell. Physiol. 2019, 234, 10080–10100.
- Welch, J.D.; Baran-Gale, J.; Perou, C.M.; Sethupathy, P.; Prins, J.F. Pseudogenes Transcribed in Breast Invasive Carcinoma Show Subtype-Specific Expression and CeRNA Potential. BMC Genom. 2015, 16, 113.
- Zhu, K.; Wang, Q.; Wang, L. Analysis of Competitive Endogenous RNA Regulatory Network of Exosomal Breast Cancer Based on ExoRBase. Evol. Bioinform. Online 2022, 18, 11769343221113286.
- Rieger, M.A.; Ebner, R.; Bell, D.R.; Kiessling, A.; Rohayem, J.; Schmitz, M.; Temme, A.; Rieber, E.P.; Weigle, B. Identification of a Novel Mammary-Restricted Cytochrome P450, CYP4Z1, with Overexpression in Breast Carcinoma. Cancer Res. 2004, 64, 2357–2364.
- Yu, W.; Chai, H.; Li, Y.; Zhao, H.; Xie, X.; Zheng, H.; Wang, C.; Wang, X.; Yang, G.; Cai, X.; et al. Increased Expression of CYP4Z1 Promotes Tumor Angiogenesis and Growth in Human Breast Cancer. Toxicol. Appl. Pharmacol. 2012, 264, 73–83.
- Zheng, L.; Li, X.; Gu, Y.; Ma, Y.; Xi, T. Pseudogene CYP4Z2P 3′UTR Promotes Angiogenesis in Breast Cancer. Biochem. Biophys. Res. Commun. 2014, 453, 545–551.
- Zheng, L.; Li, X.; Gu, Y.; Lv, X.; Xi, T. The 3′UTR of the Pseudogene CYP4Z2P Promotes Tumor Angiogenesis in Breast Cancer by Acting as a CeRNA for CYP4Z1. Breast Cancer Res. Treat. 2015, 150, 105–118, Erratum in Breast Cancer Res. Treat. 2020, 179, 521–522.
- Zheng, L.; Li, X.; Meng, X.; Chou, J.; Hu, J.; Zhang, F.; Zhang, Z.; Xing, Y.; Liu, Y.; Xi, T. Competing Endogenous RNA Networks of CYP4Z1 and Pseudogene CYP4Z2P Confer Tamoxifen Resistance in Breast Cancer. Mol. Cell. Endocrinol. 2016, 427, 133–142.
- Li, C.; Zheng, L.; Xin, Y.; Tan, Z.; Zhang, Y.; Meng, X.; Wang, Z.; Xi, T. The Competing Endogenous RNA Network of CYP4Z1 and Pseudogene CYP4Z2P Exerts an Anti-Apoptotic Function in Breast Cancer. FEBS Lett. 2017, 591, 991–1000.
- Zheng, L.; Guo, Q.; Xiang, C.; Liu, S.; Jiang, Y.; Gao, L.; Ni, H.; Wang, T.; Zhao, Q.; Liu, H.; et al. Transcriptional Factor Six2 Promotes the Competitive Endogenous RNA Network between CYP4Z1 and Pseudogene CYP4Z2P Responsible for Maintaining the Stemness of Breast Cancer Cells. J. Hematol. Oncol. 2019, 12, 23, Erratum in J. Hematol. Oncol. 2019, 12, 109.
- Ching, Y.P.; Wong, C.M.; Chan, S.F.; Leung, T.H.Y.; Ng, D.C.H.; Jin, D.Y.; Ng, I.O.L. Deleted in Liver Cancer (DLC) 2 Encodes a RhoGAP Protein with Growth Suppressor Function and Is Underexpressed in Hepatocellular Carcinoma. J. Biol. Chem. 2003, 278, 10824–10830.
- Lin, Y.; Chen, N.T.; Shih, Y.P.; Liao, Y.C.; Xue, L.; Lo, S.H. DLC2 Modulates Angiogenic Responses in Vascular Endothelial Cells by Regulating Cell Attachment and Migration. Oncogene 2010, 29, 3010–3016.
- Ullmannova, V.; Popescu, N.C. Expression Profile of the Tumor Suppressor Genes DLC-1 and DLC-2 in Solid Tumors. Int. J. Oncol. 2006, 29, 1127–1132.
- Hanna, S.; Khalil, B.; Nasrallah, A.; Saykali, B.A.; Sobh, R.; Nasser, S.; El-Sibai, M. StarD13 Is a Tumor Suppressor in Breast Cancer That Regulates Cell Motility and Invasion. Int. J. Oncol. 2014, 44, 1499–1511.
- Hu, J.; Li, X.; Guo, X.; Guo, Q.; Xiang, C.; Zhang, Z.; Xing, Y.; Xi, T.; Zheng, L. The CCR2 3′UTR Functions as a Competing Endogenous RNA to Inhibit Breast Cancer Metastasis. J. Cell Sci. 2017, 130, 3399–3413.
- Basak, P.; Leslie, H.; Dillon, R.L.; Muller, W.J.; Raouf, A.; Mowat, M.R.A. In Vivo Evidence Supporting a Metastasis Suppressor Role for Stard13 (Dlc2) in ErbB2 (Neu) Oncogene Induced Mouse Mammary Tumors. Genes Chromosomes Cancer 2018, 57, 182–191.
- Zhou, G.; Liu, X.; Xiong, B.; Sun, Y. Homeobox B4 Inhibits Breast Cancer Cell Migration by Directly Binding to StAR-Related Lipid Transfer Domain Protein 13. Oncol. Lett. 2017, 14, 4625–4632.
- Guo, X.; Xiang, C.; Zhang, Z.; Zhang, F.; Xi, T.; Zheng, L. Displacement of Bax by BMF Mediates STARD13 3′UTR-Induced Breast Cancer Cells Apoptosis in an MiRNA-Depedent Manner. Mol. Pharm. 2018, 15, 63–71.
- Liu, Y.; Chen, Y.; Zhao, Q.; Xie, T.; Xiang, C.; Guo, Q.; Zhang, W.; Zhou, Y.; Yuan, Y.; Zhang, Y.; et al. A Positive TGF-β/MiR-9 Regulatory Loop Promotes the Expansion and Activity of Tumour-Initiating Cells in Breast Cancer. Br. J. Pharmacol. 2023, 180, 2280–2297.
- Amirfallah, A.; Knutsdottir, H.; Arason, A.; Hilmarsdottir, B.; Johannsson, O.T.; Agnarsson, B.A.; Barkardottir, R.B.; Reynisdottir, I. Hsa-MiR-21-3p Associates with Breast Cancer Patient Survival and Targets Genes in Tumor Suppressive Pathways. PLoS ONE 2021, 16, e260327.
- Zheng, L.; Xiang, C.; Li, X.; Guo, Q.; Gao, L.; Ni, H.; Xia, Y.; Xi, T. STARD13-Correlated CeRNA Network-Directed Inhibition on YAP/TAZ Activity Suppresses Stemness of Breast Cancer via Co-Regulating Hippo and Rho-GTPase/F-Actin Signaling. J. Hematol. Oncol. 2018, 11, 72.
- Li, X.; Zheng, L.; Zhang, F.; Hu, J.; Chou, J.; Liu, Y.; Xing, Y.; Xi, T. STARD13-Correlated CeRNA Network Inhibits EMT and Metastasis of Breast Cancer. Oncotarget 2016, 7, 23197–23211.
- Seillier, M.; Peuget, S.; Gayet, O.; Gauthier, C.; N’Guessan, P.; Monte, M.; Carrier, A.; Iovanna, J.L.; Dusetti, N.J. TP53INP1, a Tumor Suppressor, Interacts with LC3 and ATG8-Family Proteins through the LC3-Interacting Region (LIR) and Promotes Autophagy-Dependent Cell Death. Cell Death Differ. 2012, 19, 1525–1535.
- Seux, M.; Peuget, S.; Montero, M.P.; Siret, C.; Rigot, V.; Clerc, P.; Gigoux, V.; Pellegrino, E.; Pouyet, L.; N’Guessan, P.; et al. TP53INP1 Decreases Pancreatic Cancer Cell Migration by Regulating SPARC Expression. Oncogene 2011, 30, 3049–3061.
- Zheng, L.; Li, X.; Chou, J.; Xiang, C.; Guo, Q.; Zhang, Z.; Guo, X.; Gao, L.; Xing, Y.; Xi, T. StarD13 3′-Untranslated Region Functions as a CeRNA for TP53INP1 in Prohibiting Migration and Invasion of Breast Cancer Cells by Regulating MiR-125b Activity. Eur. J. Cell Biol. 2018, 97, 23–31.
- Puthalakath, H.; Villunger, A.; O’Reilly, L.A.; Beaumont, J.G.; Coultas, L.; Cheney, R.E.; Huang, D.C.S.; Strasser, A. Bmf: A Proapoptotic BH3-Only Protein Regulated by Interaction with the Myosin V Actin Motor Complex, Activated by Anoikis. Science 2001, 293, 1829–1832.
- Li, X.; Jia, Q.; Zhou, Y.; Jiang, X.; Song, L.; Wu, Y.; Wang, A.; Chen, W.; Wang, S.; Lu, Y. Tanshinone IIA Attenuates the Stemness of Breast Cancer Cells via Targeting the MiR-125b/STARD13 Axis. Exp. Hematol. Oncol. 2022, 11, 2.
More
Information
Subjects:
Anatomy & Morphology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
417
Revisions:
2 times
(View History)
Update Date:
28 Feb 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




