
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Frejus Sodedji | -- | 1650 | 2024-02-10 06:34:47 | | | |
| 2 | Jason Zhu | Meta information modification | 1650 | 2024-02-18 03:03:53 | | |
Video Upload Options
Legumes are a source of important secondary metabolites including carotenoids, and they play a significant role in food and diet diversification and ecosystem protection. Carotenoids are the second-most abundant naturally occurring pigments on earth, synthesized by plants, which fulfill important physiological functions. The main carotenoid pigments found in the photosystems of plants include α-Carotene and β-carotene, which are further hydroxylated to produce xanthophylls (e.g., lutein and zeaxanthin). The crucial roles of carotenoids and their metabolites in photooxidative protection and photosynthesis, not to mention nutrition, vision, and cellular differentiation, make them an important class of biological pigments. In cowpeas, carotenoids are mainly present in seeds, leaves, and pods, which contribute to the antioxidant properties of this legume. Increasing the content of carotenoids in cowpeas will contribute to food and nutrition security in the tropics.
1. Breeding for Increased Carotenoid Content in Cowpea
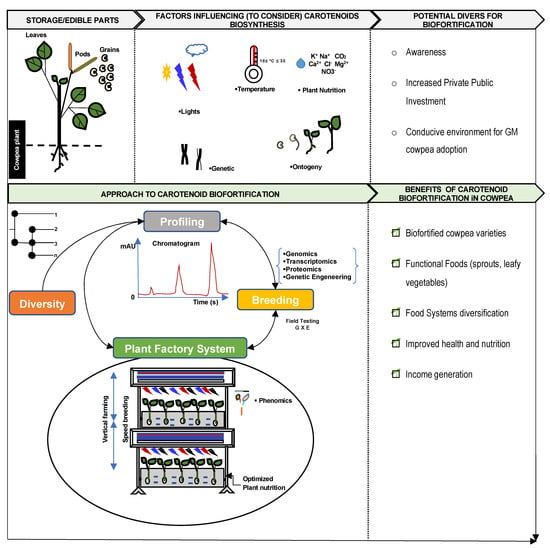
2. Harnessing the Power of Plant Factory System, Speed Breeding, and Omics
3. Genetic Engineering for Increasing Carotenoid Content in Cowpea
References
- Bouis, H.; Low, J.; McEwan, M.; Tanumihardjo, S. Biofortification: Evidence and Lessons Learned Linking Agriculture and Nutrition; FAO: Rome, Italy, 2013; pp. 1–23.
- Bouis, H.E.; Hotz, C.; McClafferty, B.; Meenakshi, J.V.; Pfeiffer, W.H. Biofortification: A new tool to reduce micronutrient malnutrition. Food Nutr. Bull. 2011, 32, S31–S40.
- Andersson, M. Progress update: Crop development of biofortified staple food crops under HarvestPlus. Afr. J. Food Agric. Nutr. Dev. 2017, 17, 11905–11935.
- Ephraim, N.; Evans, A. Advances in carotenoid increments in storage parts of African staple crops. J. Plant Breed. Crop Sci. 2019, 11, 68–79.
- Gedil, M.; Menkir, A. An Integrated Molecular and Conventional Breeding Scheme for Enhancing Genetic Gain in Maize in Africa. Front. Plant Sci. 2019, 10, 1430.
- Mahalakshmi, V.; Ng, Q.; Lawson, M.; Ortiz, R. Cowpea core collection defined by geographical, agronomical and botanical descriptors. Plant Genet. Resour. Charact. Util. 2007, 5, 113–119.
- Fatokun, C.; Girma, G.; Abberton, M.; Gedil, M.; Unachukwu, N.; Oyatomi, O.; Yusuf, M.; Rabbi, I.; Boukar, O. Genetic diversity and population structure of a mini-core subset from the world cowpea (Vigna unguiculata (L.) Walp.) germplasm collection. Sci. Rep. 2018, 8, 16035.
- Muñoz-Amatriaín, M.; Lo, S.; Herniter, I.A.; Boukar, O.; Fatokun, C.; Carvalho, M.; Castro, I.; Guo, Y.N.; Huynh, B.L.; Roberts, P.A.; et al. The UCR Minicore: A valuable resource for cowpea research and breeding. Legume Sci. 2021, 3, e95.
- Qin, J.; Shi, A.; Xiong, H.; Mou, B.; Motes, D.R.; Lu, W.; Miller, J.C.; Scheuring, D.C.; Nzaramba, M.N.; Weng, Y.; et al. Population structure analysis and association mapping of seed antioxidant content in USDA cowpea (Vigna unguiculata L. Walp.) core collection using SNPs. Can. J. Plant Sci. 2016, 96, 1026–1036.
- Stanley, L.; Yuan, Y.W. Transcriptional Regulation of Carotenoid Biosynthesis in Plants: So Many Regulators, So Little Consensus. Front. Plant Sci. 2019, 10, 1017.
- Gupta, H.S.; Hossain, F.; Muthusamy, V.; Zunjare, R.U. Marker-Assisted Breeding for Enrichment of Provitamin A in Maize. In Quality Breeding in Field Crops; Springer: Cham, Switzerland, 2019; pp. 139–157.
- Boukar, O.; Belko, N.; Chamarthi, S.; Togola, A.; Batieno, J.; Owusu, E.; Haruna, M.; Diallo, S.; Umar, M.L.; Olufajo, O.; et al. Cowpea (Vigna unguiculata): Genetics, genomics and breeding. Plant Breed. 2019, 138, 415–424.
- Lonardi, S.; Muñoz-Amatriaín, M.; Liang, Q.; Shu, S.; Wanamaker, S.I.; Lo, S.; Tanskanen, J.; Schulman, A.H.; Zhu, T.; Luo, M.C.; et al. The genome of cowpea (Vigna unguiculata Walp.). Plant J. 2019, 98, 767–782.
- Ouédraogo, J.T.; Gowda, B.S.; Jean, M.; Close, T.J.; Ehlers, J.D.; Hall, A.E.; Gillaspie, A.G.; Roberts, P.A.; Ismail, A.M.; Bruening, G.; et al. An improved genetic linkage map for cowpea (Vigna unguiculata L.) Combining AFLP, RFLP, RAPD, biochemical markers, and biological resistance traits. Genome 2002, 45, 175–188.
- Pan, L.; Wang, N.; Wu, Z.; Guo, R.; Yu, X.; Zheng, Y.; Xia, Q.; Gui, S.; Chen, C. A High Density Genetic Map Derived from RAD Sequencing and Its Application in QTL Analysis of Yield-Related Traits in Vigna unguiculata. Front. Plant Sci. 2017, 8, 1544.
- Muchero, W.; Diop, N.N.; Bhat, P.R.; Fenton, R.D.; Wanamaker, S.; Pottorff, M.; Hearne, S.; Cisse, N.; Fatokun, C.; Ehlers, J.D.; et al. A consensus genetic map of cowpea and synteny based on EST-derived SNPs. Proc. Natl. Acad. Sci. USA 2009, 106, 18159–18164.
- Adetumbi, J.A.; Akinyosoye, S.T.; Olowolafe, M.O.; Oloyede-Kamiyo, Q.O.; Agbeleye, O.A. Genetic linkage map of cowpea (Vigna unguiculata (L.) Walp.) using SNP markers. Afr. J. Biotechnol. 2016, 15, 830–834.
- Chen, H.; Wang, L.; Liu, X.; Hu, L.; Wang, S.; Cheng, X. De novo transcriptomic analysis of cowpea (Vigna unguiculata L. Walp.) for genic SSR marker development. BMC Genet. 2017, 18, 65.
- Mafakheri, K.; Bihamta, M.R.; Abbasi, A.R. Assessment of genetic diversity in cowpea (Vigna unguiculata L.) germplasm using morphological and molecular characterisation. Cogent Food Agric. 2017, 3, 1327092.
- Gupta, S.K.; Gopalakrishna, T. Development of unigene-derived SSR markers in cowpea (Vigna unguiculata) and their transferability to other Vigna species. Genome 2010, 53, 508–523.
- Diouf, M.; Diallo, S.; Abaye Badiane, F.; Diack, O.; Diouf, D. Development of new cowpea (Vigna unguiculata) mutant genotypes, analysis of their agromorphological variation, genetic diversity and population structure. Biocell 2021, 45, 345–362.
- Sharp, P.; Dong, C. Tilling for plant breeding. Methods Mol. Biol. 2014, 1145, 155–165.
- Muthoni, J.; Shimelis, H. Mating designs commonly used in plant breeding: A review. Aust. J. Crop Sci. 2020, 14, 1855–1869.
- Cazzola, F.; Bermejo, C.J.; Gatti, I.; Cointry, E. Speed breeding in pulses: An opportunity to improve the efficiency of breeding programs. Crop Pasture Sci. 2021, 72, 165–172.
- Sreekala, C.; Raghava, S.P. Exploitation of heterosis for carotenoid content in African marigold (Tagetes erecta L.) and its correlation with esterase polymorphism. Theor. Appl. Genet. 2003, 106, 771–776.
- Kandianis, C.B.; Stevens, R.; Liu, W.; Palacios, N.; Montgomery, K.; Pixley, K.; White, W.S.; Rocheford, T. Genetic architecture controlling variation in grain carotenoid composition and concentrations in two maize populations. Theor. Appl. Genet. 2013, 126, 2879–2895.
- Kozai, T.; Niu, G. Plant Factory as a Resource-Efficient Closed Plant Production System. In Plant Factory; Academic Press: Cambridge, MA, USA, 2016; pp. 69–90.
- Zhang, X.; Bian, Z.; Yuan, X.; Chen, X.; Lu, C. A review on the effects of light-emitting diode (LED) light on the nutrients of sprouts and microgreens. Trends Food Sci. Technol. 2020, 99, 203–216.
- Frede, K.; Schreiner, M.; Zrenner, R.; Graefe, J.; Baldermann, S. Carotenoid biosynthesis of pak choi (Brassica rapa ssp. chinensis) sprouts grown under different light-emitting diodes during the diurnal course. Photochem. Photobiol. Sci. 2018, 17, 1289–1300.
- Ntagkas, N.; de Vos, R.C.H.; Woltering, E.J.; Nicole, C.C.S.; Labrie, C.; Marcelis, L.F.M. Modulation of the Tomato Fruit Metabolome by LED Light. Metabolites 2020, 10, 266.
- Watson, A.; Ghosh, S.; Williams, M.J.; Cuddy, W.S.; Simmonds, J.; Rey, M.D.; Asyraf Md Hatta, M.; Hinchliffe, A.; Steed, A.; Reynolds, D.; et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 2018, 4, 23–29.
- Jahne, F.; Hahn, V.; Wurschum, T.; Leiser, W.L. Speed breeding short-day crops by LED-controlled light schemes. Theor. Appl. Genet. 2020, 133, 2335–2342.
- Dhanapal, A.P.; Ray, J.D.; Singh, S.K.; Hoyos-Villegas, V.; Smith, J.R.; Purcell, L.C.; King, C.A.; Fritschi, F.B. Association mapping of total carotenoids in diverse soybean genotypes based on leaf extracts and high-throughput canopy spectral reflectance measurements. PLoS ONE 2015, 10, e0137213.
- Chandra, A.; Bhatt, R.K. Biochemical and physiological response to salicylic acid in relation to the systemic acquired resistance. Photosynthetica 1998, 35, 255–258.
- Stoleru, V.; Burlica, R.; Mihalache, G.; Dirlau, D.; Padureanu, S.; Teliban, G.C.; Astanei, D.; Cojocaru, A.; Beniuga, O.; Patras, A. Plant growth promotion effect of plasma activated water on Lactuca sativa L. cultivated in two different volumes of substrate. Sci. Rep. 2020, 10, 20920.
- Hempel, J.; Müller-Maatsch, J.; Carle, R.; Schweiggert, R.M. Non-destructive approach for the characterization of the in situ carotenoid deposition in gac fruit aril. J. Food Compos. Anal. 2018, 65, 16–22.
- Akpolat, H.; Barineau, M.; Jackson, K.A.; Akpolat, M.Z.; Francis, D.M.; Chen, Y.J.; Rodriguez-Saona, L.E. High-throughput phenotyping approach for screening major carotenoids of tomato by handheld raman spectroscopy using chemometric methods. Sensors 2020, 20, 3723.
- Araus, J.L.; Kefauver, S.C.; Zaman-Allah, M.; Olsen, M.S.; Cairns, J.E. Translating High-Throughput Phenotyping into Genetic Gain. Trends Plant Sci. 2018, 23, 451–466.
- Varshney, R.K.; Bohra, A.; Yu, J.; Graner, A.; Zhang, Q.; Sorrells, M.E. Designing Future Crops: Genomics-Assisted Breeding Comes of Age. Trends Plant Sci. 2021, 26, 631–649.
- Zheng, X.; Kuijer, H.N.J.; Al-Babili, S. Carotenoid Biofortification of Crops in the CRISPR Era. Trends Biotechnol. 2020, 39, 857–860.
- Fernández-Marín, B.; Milla, R.; Martín-Robles, N.; Arc, E.; Kranner, I.; Becerril, J.M.; García-Plazaola, J.I. Side-effects of domestication: Cultivated legume seeds contain similar tocopherols and fatty acids but less carotenoids than their wild counterparts. BMC Plant Biol. 2014, 14, 1599.
- Jha, A.B.; Warkentin, T.D. Biofortification of pulse crops: Status and future perspectives. Plants 2020, 9, 73.
- Mudziwapasi, R.; Ndudzo, A.; Nyamusamba, R.P.; Jomane, F.N.; Mutengwa, T.T.; Maphosa, M. Unlocking the potential of CRISPR technology for improving livelihoods in Africa. Biotechnol. Genet. Eng. Rev. 2018, 34, 198–215.
- Wada, N.; Ueta, R.; Osakabe, Y.; Osakabe, K. Precision genome editing in plants: State-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol. 2020, 20, 234.
- Romer, S.; Lubeck, J.; Kauder, F.; Steiger, S.; Adomat, C.; Sandmann, G. Genetic engineering of a zeaxanthin-rich potato by antisense inactivation and co-suppression of carotenoid epoxidation. Metab. Eng. 2002, 4, 263–272.
- Sandmann, G. Genetic manipulation of carotenoid biosynthesis: Strategies, problems and achievements. Trends Plant Sci. 2001, 6, 14–17.
- Mishiba, K.I.; Nishida, K.; Inoue, N.; Fujiwara, T.; Teranishi, S.; Iwata, Y.; Takeda, S.; Koizumi, N. Genetic engineering of eggplant accumulating beta-carotene in fruit. Plant Cell Rep. 2020, 39, 1029–1039.
- Badejo, A.A. Elevated carotenoids in staple crops: The biosynthesis, challenges and measures for target delivery. J. Genet. Eng. Biotechnol. 2018, 16, 553–562.
- Komen, J.; Tripathi, L.; Mkoko, B.; Ofosu, D.O.; Oloka, H.; Wangari, D. Biosafety Regulatory Reviews and Leeway to Operate: Case Studies from Sub-Sahara Africa. Front. Plant Sci. 2020, 11, 130.




