
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Kefeng Li | -- | 3078 | 2024-02-01 05:03:33 | | | |
| 2 | Lindsay Dong | Meta information modification | 3078 | 2024-02-02 07:33:43 | | |
Video Upload Options
The interconnectivity of advanced biological systems is essential for their proper functioning. In modern connectomics, biological entities such as proteins, genes, RNA, DNA, and metabolites are often represented as nodes, while the physical, biochemical, or functional interactions between them are represented as edges. Among these entities, metabolites are particularly significant as they exhibit a closer relationship to an organism’s phenotype compared to genes or proteins. Moreover, the metabolome has the ability to amplify small proteomic and transcriptomic changes, even those from minor genomic changes. Metabolic networks, which consist of complex systems comprising hundreds of metabolites and their interactions, play a critical role in biological research by mediating energy conversion and chemical reactions within cells.
1. Introduction
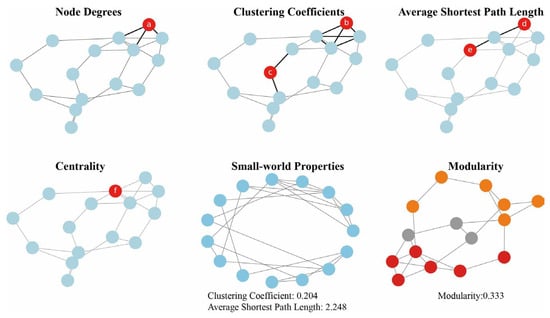
2. Construction Methods of Metabolic Networks
| Metabolic Network | Method/Model | Language | Source |
|---|---|---|---|
| Correlation-based | Pearson correlation And Spearman rank correlation | Python | https://github.com/aishapectyo/Correlations-Pearson-Spearman (accessed on 28 November 2023) |
| Distance correlation [20] | Python | https://github.com/vnmabus/dcor (accessed on 28 November 2023) | |
| Gaussian graphical model | R | https://github.com/donaldRwilliams/BGGM (accessed on 28 November 2023) | |
| Causal-based | Causal inference model [21] | Python | https://github.com/BiomedSciAI/causallib (accessed on 28 November 2023) |
| Structural equation model | R | https://github.com/yrosseel/lavaan (accessed on 28 November 2023) | |
| Dynamic causal model | Python | https://github.com/tmdemelo/pydcm (accessed on 28 November 2023) | |
| Pathway-based | Pathway | Python | https://github.com/iseekwonderful/PyPathway (accessed on 28 November 2023) |
| Chemical structure similarity-based | Chemical structure similarity | Python | https://github.com/labsyspharm/lsp-cheminformatics (accessed on 28 November 2023) |
2.1. Correlation-Based Metabolic Network
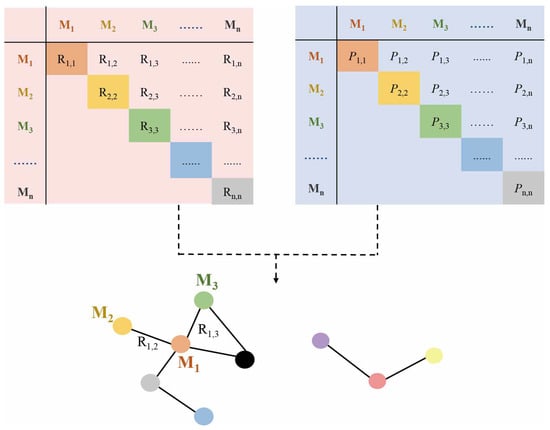
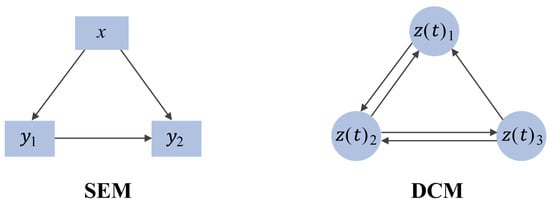
2.2. Causal-Based Metabolic Network
2.3. Pathway-Based Metabolic Network
2.4. Metabolic Network Based on Chemical Structure Similarity
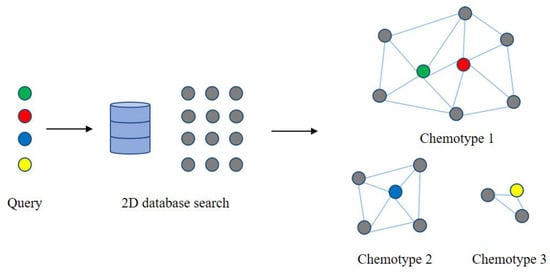
3. Application of Metabolic Network
3.1. Metabolic Networks in Disease Mechanisms
3.2. Metabolic Networks in Disease Prediction and Diagnosis
3.3. Drug Discovery and Disease Treatment
4. Conclusions
References
- Assenov, Y.; Ramirez, F.; Schelhorn, S.E.; Lengauer, T.; Albrecht, M. Computing topological parameters of biological networks. Bioinformatics 2008, 24, 282–284.
- Muzio, G.; O’Bray, L.; Borgwardt, K. Biological network analysis with deep learning. Brief. Bioinform. 2021, 22, 1515–1530.
- Klein, C.; Marino, A.; Sagot, M.F.; Vieira Milreu, P.; Brilli, M. Structural and dynamical analysis of biological networks. Brief. Funct. Genom. 2012, 11, 420–433.
- Mengiste, S.A.; Aertsen, A.; Kumar, A. Relevance of network topology for the dynamics of biological neuronal networks. bioRxiv 2021.
- May, R.M. Network structure and the biology of populations. Trends Ecol. Evol. 2006, 21, 394–399.
- Melo, D.; Porto, A.; Cheverud, J.M.; Marroig, G. Modularity: Genes, development and evolution. Annu. Rev. Ecol. Evol. Syst. 2016, 47, 463–486.
- Lorenz, D.M.; Jeng, A.; Deem, M.W. The emergence of modularity in biological systems. Phys. Life Rev. 2011, 8, 129–160.
- Lee, U.; Mashour, G.A. Role of Network Science in the Study of Anesthetic State Transitions. Anesthesiology 2018, 129, 1029–1044.
- De Las Rivas, J.; Fontanillo, C. Protein-protein interactions essentials: Key concepts to building and analyzing interactome networks. PLoS Comput. Biol. 2010, 6, e1000807.
- Wu, M.; Su, R.Q.; Li, X.; Ellis, T.; Lai, Y.C.; Wang, X. Engineering of regulated stochastic cell fate determination. Proc. Natl. Acad. Sci. USA 2013, 110, 10610–10615.
- Folger, O.; Jerby, L.; Frezza, C.; Gottlieb, E.; Ruppin, E.; Shlomi, T. Predicting selective drug targets in cancer through metabolic networks. Mol. Syst. Biol. 2011, 7, 501.
- Park, H.J.; Friston, K. Structural and functional brain networks: From connections to cognition. Science 2013, 342, 1238411.
- Girvan, M.; Newman, M.E. Community structure in social and biological networks. Proc. Natl. Acad. Sci. USA 2002, 99, 7821–7826.
- Amara, A.; Frainay, C.; Jourdan, F.; Naake, T.; Neumann, S.; Novoa-Del-Toro, E.M.; Salek, R.M.; Salzer, L.; Scharfenberg, S.; Witting, M. Networks and Graphs Discovery in Metabolomics Data Analysis and Interpretation. Front. Mol. Biosci. 2022, 9, 841373.
- Yazdani, A.; Yazdani, A.; Mendez-Giraldez, R.; Samiei, A.; Kosorok, M.R.; Schaid, D.J. From classical mendelian randomization to causal networks for systematic integration of multi-omics. Front. Genet. 2022, 13, 990486.
- Ness, R.O.; Sachs, K.; Vitek, O. From Correlation to Causality: Statistical Approaches to Learning Regulatory Relationships in Large-Scale Biomolecular Investigations. J. Proteome Res. 2016, 15, 683–690.
- Rohrer, J.M. Thinking Clearly About Correlations and Causation: Graphical Causal Models for Observational Data. Adv. Methods Pract. Psychol. Sci. 2018, 1, 27–42.
- Hattori, M.; Okuno, Y.; Goto, S.; Kanehisa, M. Development of a chemical structure comparison method for integrated analysis of chemical and genomic information in the metabolic pathways. J. Am. Chem. Soc. 2003, 125, 11853–11865.
- Holliday, G.L.; Andreini, C.; Fischer, J.D.; Rahman, S.A.; Almonacid, D.E.; Williams, S.T.; Pearson, W.R. MACiE: Exploring the diversity of biochemical reactions. Nucleic Acids Res. 2012, 40, D783–D789.
- Ramos-Carreño, C.; Torrecilla, J.L. dcor: Distance correlation and energy statistics in Python. SoftwareX 2023, 22, 101326.
- Shimoni, Y.; Karavani, E.; Ravid, S.; Bak, P.; Ng, T.H.M.; Alford, S.H.; Meade, D.; Goldschmidt, Y. An Evaluation Toolkit to Guide Model Selection and Cohort Definition in Causal Inference. arXiv 2019, arXiv:1906.00442.
- Batushansky, A.; Toubiana, D.; Fait, A. Correlation-Based Network Generation, Visualization, and Analysis as a Powerful Tool in Biological Studies: A Case Study in Cancer Cell Metabolism. Biomed. Res. Int. 2016, 2016, 8313272.
- Nishihara, R.; Glass, K.; Mima, K.; Hamada, T.; Nowak, J.A.; Qian, Z.R.; Kraft, P.; Giovannucci, E.L.; Fuchs, C.S.; Chan, A.T.; et al. Biomarker correlation network in colorectal carcinoma by tumor anatomic location. BMC Bioinform. 2017, 18, 304.
- Kotze, H.L.; Armitage, E.G.; Sharkey, K.J.; Allwood, J.W.; Dunn, W.B.; Williams, K.J.; Goodacre, R. A novel untargeted metabolomics correlation-based network analysis incorporating human metabolic reconstructions. BMC Syst. Biol. 2013, 7, 107.
- Chen, P.Y.; Cripps, A.W.; West, N.P.; Cox, A.J.; Zhang, P. A correlation-based network for biomarker discovery in obesity with metabolic syndrome. BMC Bioinform. 2019, 20, 477.
- Jahagirdar, S.; Suarez-Diez, M.; Saccenti, E. Simulation and Reconstruction of Metabolite-Metabolite Association Networks Using a Metabolic Dynamic Model and Correlation Based Algorithms. J. Proteome Res. 2019, 18, 1099–1113.
- de Siqueira Santos, S.; Takahashi, D.Y.; Nakata, A.; Fujita, A. A comparative study of statistical methods used to identify dependencies between gene expression signals. Brief. Bioinform. 2014, 15, 906–918.
- Aguilera-Mendoza, L.; Marrero-Ponce, Y.; Garcia-Jacas, C.R.; Chavez, E.; Beltran, J.A.; Guillen-Ramirez, H.A.; Brizuela, C.A. Automatic construction of molecular similarity networks for visual graph mining in chemical space of bioactive peptides: An unsupervised learning approach. Sci. Rep. 2020, 10, 18074.
- Kumari, S.; Nie, J.; Chen, H.S.; Ma, H.; Stewart, R.; Li, X.; Lu, M.Z.; Taylor, W.M.; Wei, H. Evaluation of gene association methods for coexpression network construction and biological knowledge discovery. PLoS ONE 2012, 7, e50411.
- Allen, E.; Moing, A.; Ebbels, T.M.; Maucourt, M.; Tomos, A.D.; Rolin, D.; Hooks, M.A. Correlation Network Analysis reveals a sequential reorganization of metabolic and transcriptional states during germination and gene-metabolite relationships in developing seedlings of Arabidopsis. BMC Syst. Biol. 2010, 4, 62.
- Dyrba, M.; Mohammadi, R.; Grothe, M.J.; Kirste, T.; Teipel, S.J. Gaussian Graphical Models Reveal Inter-Modal and Inter-Regional Conditional Dependencies of Brain Alterations in Alzheimer’s Disease. Front. Aging Neurosci. 2020, 12, 99.
- Krumsiek, J.; Suhre, K.; Illig, T.; Adamski, J.; Theis, F.J. Gaussian graphical modeling reconstructs pathway reactions from high-throughput metabolomics data. BMC Syst. Biol. 2011, 5, 21.
- Hackett, S.R.; Baltz, E.A.; Coram, M.; Wranik, B.J.; Kim, G.; Baker, A.; Fan, M.; Hendrickson, D.G.; Berndl, M.; McIsaac, R.S. Learning causal networks using inducible transcription factors and transcriptome-wide time series. Mol. Syst. Biol. 2020, 16, e9174.
- Perfetto, L.; Briganti, L.; Calderone, A.; Perpetuini, A.C.; Lannuccelli, M.; Langone, F.; Licata, L.; Marinkovic, M.; Mattioni, A.; Pavlidou, T.; et al. SIGNOR: A database of causal relationships between biological entities. Nucleic Acids Res. 2016, 44, D548–D554.
- Yao, L.Y.; Chu, Z.X.; Li, S.; Li, Y.L.; Gao, J.; Zhang, A.D. A Survey on Causal Inference. ACM Trans. Knowl. Discov. Data 2021, 15, 74.
- Nogueira, A.R.; Pugnana, A.; Ruggieri, S.; Pedreschi, D.; Gama, J. Methods and tools for causal discovery and causal inference. Wiley Interdiscip. Rev.-Data Min. Knowl. Discov. 2022, 12, e1449.
- Rosa, G.J.; Valente, B.D.; de los Campos, G.; Wu, X.L.; Gianola, D.; Silva, M.A. Inferring causal phenotype networks using structural equation models. Genet. Sel. Evol. 2011, 43, 6.
- Friston, K. Dynamic causal modeling and Granger causality Comments on: The identification of interacting networks in the brain using fMRI: Model selection, causality and deconvolution. Neuroimage 2011, 58, 303–305, author reply 310–311.
- Peters, J.; Janzing, D.; Schölkopf, B. Causal inference on time series using restricted structural equation models. Adv. Neural Inf. Process. Syst. 2013, 26, 154–162.
- Rose, N.; Wagner, W.; Mayer, A.; Nagengast, B. Model-Based Manifest and Latent Composite Scores in Structural Equation Models. Collabra Psychol. 2019, 5, 9.
- Bollen, K.A.; Hoyle, R.H. Latent variables in structural equation modeling. In Handbook of Structural Equation Modeling; Hoyle, R.H., Ed.; Guilford Press: New York, NY, USA, 2012; pp. 56–67.
- Yuan, K.-H.; Kouros, C.D.; Kelley, K. Diagnosis for covariance structure models by analyzing the path. Struct. Equ. Model. A Multidiscip. J. 2008, 15, 564–602.
- Friston, K.J. Functional and effective connectivity: A review. Brain Connect. 2011, 1, 13–36.
- Wang, J.; Sun, Y.; Teng, S.; Li, K. Prediction of sepsis mortality using metabolite biomarkers in the blood: A meta-analysis of death-related pathways and prospective validation. BMC Med. 2020, 18, 83.
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361.
- Caspi, R.; Billington, R.; Ferrer, L.; Foerster, H.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Mueller, L.A.; et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016, 44, D471–D480.
- King, Z.A.; Lu, J.; Drager, A.; Miller, P.; Federowicz, S.; Lerman, J.A.; Ebrahim, A.; Palsson, B.O.; Lewis, N.E. BiGG Models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 2016, 44, D515–D522.
- Henry, C.S.; DeJongh, M.; Best, A.A.; Frybarger, P.M.; Linsay, B.; Stevens, R.L. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 2010, 28, 977–982.
- Wang, L.; Dash, S.; Ng, C.Y.; Maranas, C.D. A review of computational tools for design and reconstruction of metabolic pathways. Synth. Syst. Biotechnol. 2017, 2, 243–252.
- Yamanishi, Y.; Araki, M.; Gutteridge, A.; Honda, W.; Kanehisa, M. Prediction of drug-target interaction networks from the integration of chemical and genomic spaces. Bioinformatics 2008, 24, i232–i240.
- Lo, Y.-C.; Torres, J.Z. Chemical similarity networks for drug discovery. Spec. Top. Drug Discov. 2016, 1, 53–70.
- Wale, N.; Watson, I.A.; Karypis, G. Comparison of descriptor spaces for chemical compound retrieval and classification. Knowl. Inf. Syst. 2008, 14, 347–375.
- Hu, G.; Kuang, G.; Xiao, W.; Li, W.; Liu, G.; Tang, Y. Performance evaluation of 2D fingerprint and 3D shape similarity methods in virtual screening. J. Chem. Inf. Model. 2012, 52, 1103–1113.
- Willett, P. Similarity searching using 2D structural fingerprints. Methods Mol. Biol. 2011, 672, 133–158.
- Bajusz, D.; Rácz, A.; Héberger, K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J. Cheminformatics 2015, 7, 20.
- Lo, Y.C.; Rensi, S.E.; Torng, W.; Altman, R.B. Machine learning in chemoinformatics and drug discovery. Drug Discov. Today 2018, 23, 1538–1546.
- Willett, P. Similarity-based data mining in files of two-dimensional chemical structures using fingerprint measures of molecular resemblance. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2011, 1, 241–251.
- Ma, H.; Goryanin, I. Human metabolic network reconstruction and its impact on drug discovery and development. Drug Discov. Today 2008, 13, 402–408.
- Li, K.; Bertrand, K.; Naviaux, J.C.; Monk, J.M.; Wells, A.; Wang, L.; Lingampelly, S.S.; Naviaux, R.K.; Chambers, C. Metabolomic and exposomic biomarkers of risk of future neurodevelopmental delay in human milk. Pediatr. Res. 2023, 93, 1710–1720.
- Judge, A.; Dodd, M.S. Metabolism. Essays Biochem. 2020, 64, 607–647.
- Dräger, A.; Planatscher, H. Metabolic Networks. In Encyclopedia of Systems Biology; Dubitzky, W., Wolkenhauer, O., Cho, K.-H., Yokota, H., Eds.; Springer: New York, NY, USA, 2013; pp. 1249–1251.
- Waller, T.C.; Berg, J.A.; Lex, A.; Chapman, B.E.; Rutter, J. Compartment and hub definitions tune metabolic networks for metabolomic interpretations. GigaScience 2020, 9, giz137.
- Theorell, A.; Stelling, J. Metabolic networks, microbial consortia, and analogies to smart grids. Proc. IEEE 2022, 110, 541–556.
- Burke, P.E.P.; Campos, C.B.L.; Costa, L.D.F.; Quiles, M.G. A biochemical network modeling of a whole-cell. Sci. Rep. 2020, 10, 13303.
- Frainay, C.; Jourdan, F. Computational methods to identify metabolic sub-networks based on metabolomic profiles. Brief. Bioinform. 2017, 18, 43–56.
- Väremo, L.; Nookaew, I.; Nielsen, J. Novel insights into obesity and diabetes through genome-scale metabolic modeling. Front. Physiol. 2013, 4, 92.
- Faust, K.; Croes, D.; van Helden, J. Prediction of metabolic pathways from genome-scale metabolic networks. Biosystems 2011, 105, 109–121.
- Hecht, F. On the origins of cancer genetics and cytogenetics. Cancer Genet. Cytogenet. 1987, 29, 187–190.
- Bintener, T.; Pacheco, M.P.; Sauter, T. Towards the routine use of in silico screenings for drug discovery using metabolic modelling. Biochem. Soc. Trans. 2020, 48, 955–969.
- Beguerisse-Díaz, M.; Bosque, G.; Oyarzún, D.; Picó, J.; Barahona, M. Flux-dependent graphs for metabolic networks. NPJ Syst. Biol. Appl. 2018, 4, 32.
- Tomar, N.; De, R.K. Comparing methods for metabolic network analysis and an application to metabolic engineering. Gene 2013, 521, 1–14.
- Zelezniak, A.; Pers, T.H.; Soares, S.; Patti, M.E.; Patil, K.R. Metabolic network topology reveals transcriptional regulatory signatures of type 2 diabetes. PLoS Comput. Biol. 2010, 6, e1000729.
- Zimmet, P.; Alberti, K.G.; Shaw, J. Global and societal implications of the diabetes epidemic. Nature 2001, 414, 782–787.
- Hameed, I.; Masoodi, S.R.; Mir, S.A.; Nabi, M.; Ghazanfar, K.; Ganai, B.A. Type 2 diabetes mellitus: From a metabolic disorder to an inflammatory condition. World J. Diabetes 2015, 6, 598–612.
- Lee, M.H.; Hwang, Y.H.; Yun, C.S.; Han, B.S.; Kim, D.Y. Altered small-world property of a dynamic metabolic network in murine left hippocampus after exposure to acute stress. Sci. Rep. 2022, 12, 3885.
- Wei, P.J.; Ma, W.; Li, Y.; Su, Y. Disease biomarker identification based on sample network optimization. Methods 2023, 213, 42–49.
- Hu, Y.; Zhao, T.; Zhang, N.; Zang, T.; Zhang, J.; Cheng, L. Identifying diseases-related metabolites using random walk. BMC Bioinform. 2018, 19, 116.
- Lei, X.; Tie, J. Prediction of disease-related metabolites using bi-random walks. PLoS ONE 2019, 14, e0225380.
- Baumgartner, C.; Spath-Blass, V.; Niederkofler, V.; Bergmoser, K.; Langthaler, S.; Lassnig, A.; Rienmüller, T.; Baumgartner, D.; Asnani, A.; Gerszten, R.E. A novel network-based approach for discovering dynamic metabolic biomarkers in cardiovascular disease. PLoS ONE 2018, 13, e0208953.
- Kerk, S.A.; Papagiannakopoulos, T.; Shah, Y.M.; Lyssiotis, C.A. Metabolic networks in mutant KRAS-driven tumours: Tissue specificities and the microenvironment. Nat. Rev. Cancer 2021, 21, 510–525.
- Hiller, K.; Metallo, C.M. Profiling metabolic networks to study cancer metabolism. Curr. Opin. Biotechnol. 2013, 24, 60–68.
- Kell, D.B.; Goodacre, R. Metabolomics and systems pharmacology: Why and how to model the human metabolic network for drug discovery. Drug Discov. Today 2014, 19, 171–182.
- Shaked, I.; Oberhardt, M.A.; Atias, N.; Sharan, R.; Ruppin, E. Metabolic Network Prediction of Drug Side Effects. Cell Syst. 2016, 2, 209–213.
- Karta, J.; Bossicard, Y.; Kotzamanis, K.; Dolznig, H.; Letellier, E. Mapping the Metabolic Networks of Tumor Cells and Cancer-Associated Fibroblasts. Cells 2021, 10, 304.
- Fouladiha, H.; Marashi, S.A. Biomedical applications of cell- and tissue-specific metabolic network models. J. Biomed. Inform. 2017, 68, 35–49.
- Lewis, N.E.; Abdel-Haleem, A.M. The evolution of genome-scale models of cancer metabolism. Front. Physiol. 2013, 4, 237.
- Di Filippo, M.; Colombo, R.; Damiani, C.; Pescini, D.; Gaglio, D.; Vanoni, M.; Alberghina, L.; Mauri, G. Zooming-in on cancer metabolic rewiring with tissue specific constraint-based models. Comput. Biol. Chem. 2016, 62, 60–69.
- Agren, R.; Bordel, S.; Mardinoglu, A.; Pornputtapong, N.; Nookaew, I.; Nielsen, J. Reconstruction of genome-scale active metabolic networks for 69 human cell types and 16 cancer types using INIT. PLoS Comput. Biol. 2012, 8, e1002518.




