
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mattias Collin | -- | 4105 | 2024-01-26 11:09:18 | | | |
| 2 | Jason Zhu | -2 word(s) | 4103 | 2024-01-29 03:28:37 | | |
Video Upload Options
Streptococcus pyogenes, or Group A Streptococcus, is an exclusively human pathogen that causes a wide variety of diseases ranging from mild throat and skin infections to severe invasive disease. The pathogenesis of S. pyogenes infection has been extensively studied, but the pathophysiology, especially of the more severe infections, is still somewhat elusive. One key feature of S. pyogenes is the expression of secreted, surface-associated, and intracellular enzymes that directly or indirectly affect both the innate and adaptive host immune systems. Undoubtedly, S. pyogenes is one of the major bacterial sources for immunomodulating enzymes. Major targets for these enzymes are immunoglobulins that are destroyed or modified through proteolysis or glycan hydrolysis.
1. Immunoglobulin Degrading and Modifying Enzymes
1.1. Immunoglobulin Cysteine Proteinases
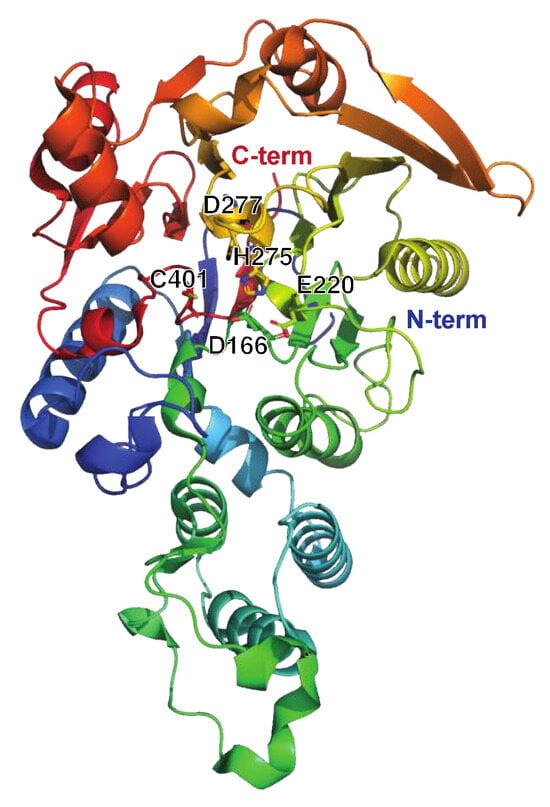
The Broad-Spectrum Cysteine Proteinase SpeB
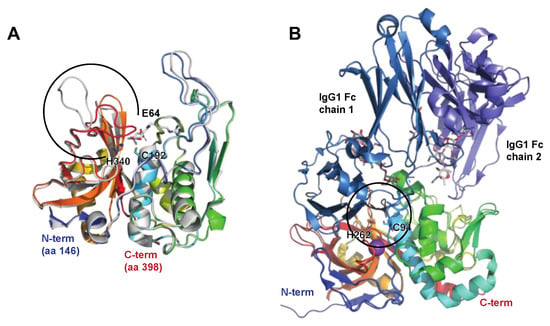
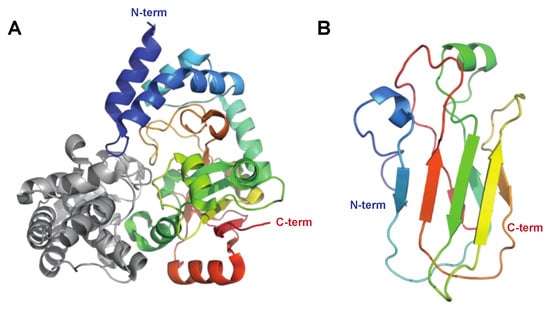
The Immunoglobulin G-Degrading Cysteine Proteinases IdeS/Mac-1/Mac-2
1.2. Immunoglobulin Glycan Hydrolases
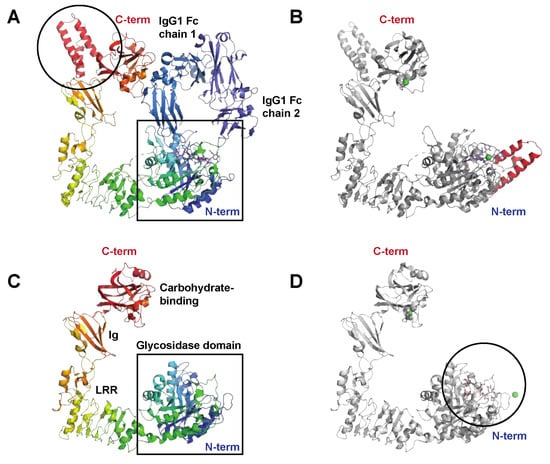
The Endo-β-N-acetylglucosaminidase EndoS
The Endo-β-N-acetylglucosaminidase EndoS2
2. Enzymes Interfering with Innate Immunity
2.1. Complement and Antimicrobial Peptide Degrading Enzymes
The Broad-Spectrum Cysteine Proteinase SpeB
The Serine Endopeptidase C5a, ScpA
2.2. Chemokine, Cytokine, and Kinin Active Enzymes
SpeB Activity on Immunologically Active Peptides
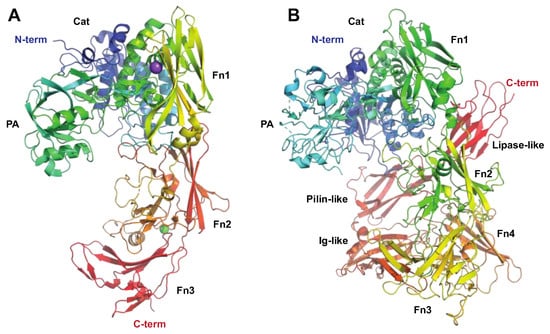
The Serine Proteinase SpyCEP
3. Enzymes Acting on Chromatin and Cellular Processes
3.1. Enzymes Acting on DNA and Nucleotides
The Streptococcal Nuclease Sda1
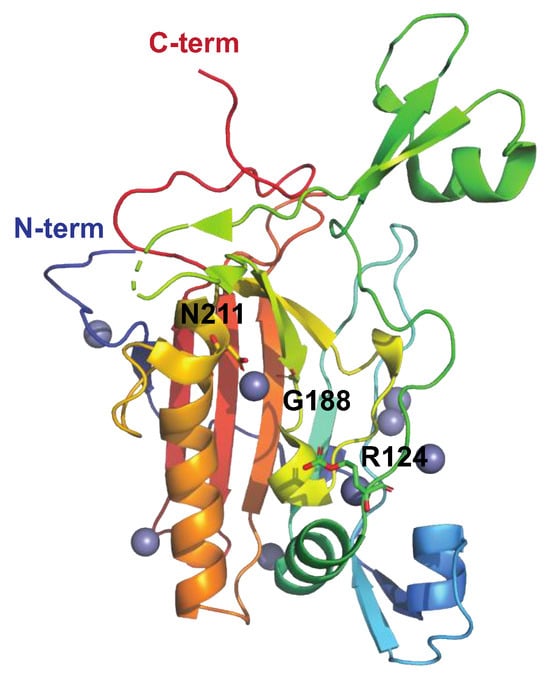
The Streptococcus pyogenes Nuclease A, SpnA
The Streptococcal 5′-Nucleotidase A, S5nA
The Streptococcal NAD(+)-Glycohydrolase, NADase
The C-di-AMP Synthase DacA
3.2. Enzymatic Effects on Pyroptosis and Autophagy
SpeB Activity on Gasdermins
SpeB Activity on Ubiquitin-Binding Proteins
2.3.3. Enzymes Acting on Other Cellular Processes
The Streptococcal Arginine Deiminase SAGP
References
- Collin, M.; Kilian, M. Bacterial modulation of Fc effector functions. In Antibody Fc; Elsevier: Amsterdam, The Netherlands, 2014; pp. 317–332.
- Walker, M.J.; Barnett, T.C.; McArthur, J.D.; Cole, J.N.; Gillen, C.M.; Henningham, A.; Sriprakash, K.S.; Sanderson-Smith, M.L.; Nizet, V. Disease manifestations and pathogenic mechanisms of Group A Streptococcus. Clin. Microbiol. Rev. 2014, 27, 264–301.
- Nelson, D.C.; Garbe, J.; Collin, M. Cysteine proteinase SpeB from Streptococcus pyogenes—A potent modifier of immunologically important host and bacterial proteins. Biol. Chem. 2011, 392, 1077–1088.
- Carroll, R.K.; Musser, J.M. From transcription to activation: How group A streptococcus, the flesh-eating pathogen, regulates SpeB cysteine protease production. Mol. Microbiol. 2011, 81, 588–601.
- Collin, M.; Olsén, A. EndoS, a novel secreted protein from Streptococcus pyogenes with endoglycosidase activity on human IgG. EMBO J. 2001, 20, 3046–3055.
- Collin, M.; Olsén, A. Effect of SpeB and EndoS from Streptococcus pyogenes on human immunoglobulins. Infect. Immun. 2001, 69, 7187–7189.
- Eriksson, A.; Norgren, M. Cleavage of antigen-bound immunoglobulin G by SpeB contributes to streptococcal persistence in opsonizing blood. Infect. Immun. 2003, 71, 211–217.
- Collin, M.; Svensson, M.D.; Sjöholm, A.G.; Jensenius, J.C.; Sjöbring, U.; Olsén, A. EndoS and SpeB from Streptococcus pyogenes inhibit immunoglobulin-mediated opsonophagocytosis. Infect. Immun. 2002, 70, 6646–6651.
- Persson, H.; Vindebro, R.; von Pawel-Rammingen, U. The streptococcal cysteine protease SpeB is not a natural immunoglobulin-cleaving enzyme. Infect. Immun. 2013, 81, 2236–2241.
- Lei, B.; DeLeo, F.R.; Hoe, N.P.; Graham, M.R.; Mackie, S.M.; Cole, R.L.; Liu, M.; Hill, H.R.; Low, D.E.; Federle, M.J.; et al. Evasion of human innate and acquired immunity by a bacterial homolog of CD11b that inhibits opsonophagocytosis. Nat. Med. 2001, 7, 1298–1305.
- von Pawel-Rammingen, U.; Johansson, B.P.; Björck, L. IdeS, a novel streptococcal cysteine proteinase with unique specificity for immunoglobulin G. EMBO J. 2002, 21, 1607–1615.
- Agniswamy, J.; Lei, B.; Musser, J.M.; Sun, P.D. Insight of host immune evasion mediated by two variants of group A Streptococcus Mac protein. J. Biol. Chem. 2004, 279, 52789–52796.
- Söderberg, J.J.; Engström, P.; von Pawel-Rammingen, U. The intrinsic immunoglobulin g endopeptidase activity of streptococcal Mac-2 proteins implies a unique role for the enzymatically impaired Mac-2 protein of M28 serotype strains. Infect. Immun. 2008, 76, 2183–2188.
- Vincents, B.; von Pawel-Rammingen, U.; Björck, L.; Abrahamson, M. Enzymatic characterization of the streptococcal endopeptidase, IdeS, reveals that it is a cysteine protease with strict specificity for IgG cleavage due to exosite binding. Biochemistry 2004, 43, 15540–15549.
- Söderberg, J.J.; von Pawel-Rammingen, U. The streptococcal protease IdeS modulates bacterial IgGFc binding and generates 1/2Fc fragments with the ability to prime polymorphonuclear leucocytes. Mol. Immunol. 2008, 45, 3347–3353.
- Wenig, K.; Chatwell, L.; von Pawel-Rammingen, U.; Björck, L.; Huber, R.; Sondermann, P. Structure of the streptococcal endopeptidase IdeS, a cysteine proteinase with strict specificity for IgG. Proc. Natl. Acad. Sci. USA 2004, 101, 17371–17376.
- Sudol, A.S.L.; Butler, J.; Ivory, D.P.; Tews, I.; Crispin, M. Extensive substrate recognition by the streptococcal antibody-degrading enzymes IdeS and EndoS. Nat. Commun. 2022, 13, 7801.
- Dardenne, L.E.; Werneck, A.S.; de Oliveira Neto, M.; Bisch, P.M. Electrostatic properties in the catalytic site of papain: A possible regulatory mechanism for the reactivity of the ion pair. Proteins 2003, 52, 236–253.
- González-Páez, G.E.; Wolan, D.W. Ultrahigh and high resolution structures and mutational analysis of monomeric Streptococcus pyogenes SpeB reveal a functional role for the glycine-rich C-terminal loop. J. Biol. Chem. 2012, 287, 24412–24426.
- Trimble, R.B.; Tarentino, A.L. Identification of distinct endoglycosidase (endo) activities in Flavobacterium meningosepticum: Endo F1, Endo F2, and Endo F3. Endo F1 and Endo H hydrolyze only high mannose and hybrid glycans. J. Biol. Chem. 1991, 266, 1646–1651.
- Sjögren, J.; Cosgrave, E.F.J.; Allhorn, M.; Nordgren, M.; Björk, S.; Olsson, F.; Fredriksson, S.; Collin, M. EndoS and EndoS2 hydrolyze Fc-glycans on therapeutic antibodies with different glycoform selectivity and can be used for rapid quantification of high-mannose glycans. Glycobiology 2015, 25, 1053–1063.
- Du, J.J.; Klontz, E.H.; Guerin, M.E.; Trastoy, B.; Sundberg, E.J. Structural insights into the mechanisms and specificities of IgG-active endoglycosidases. Glycobiology 2019, 30, 268–279.
- Klontz, E.H.; Trastoy, B.; Deredge, D.; Fields, J.K.; Li, C.; Orwenyo, J.; Marina, A.; Beadenkopf, R.; Günther, S.; Flores, J.; et al. Molecular basis of broad spectrum N-Glycan specificity and processing of therapeutic IgG monoclonal antibodies by Endoglycosidase S2. ACS Cent. Sci. 2019, 5, 524–538.
- Trastoy, B.; Lomino, J.V.; Pierce, B.G.; Carter, L.G.; Günther, S.; Giddens, J.P.; Snyder, G.A.; Weiss, T.M.; Weng, Z.; Wang, L.-X.; et al. Crystal structure of Streptococcus pyogenes EndoS, an immunomodulatory endoglycosidase specific for human IgG antibodies. Proc. Natl. Acad. Sci. USA 2014, 111, 6714–6719.
- Sjögren, J.; Struwe, W.B.; Cosgrave, E.F.J.; Rudd, P.M.; Stervander, M.; Allhorn, M.; Hollands, A.; Nizet, V.; Collin, M. EndoS2 is a unique and conserved enzyme of serotype M49 group A Streptococcus that hydrolyses N-linked glycans on IgG and α1-acid glycoprotein. Biochem. J. 2013, 455, 107–118.
- Sudol, A.S.L.; Tews, I.; Crispin, M. Bespoke conformation and antibody recognition distinguishes the streptococcal immune evasion factors EndoS and EndoS2. bioRxiv 2023.
- Trastoy, B.; Klontz, E.; Orwenyo, J.; Marina, A.; Wang, L.-X.; Sundberg, E.J.; Guerin, M.E. Structural basis for the recognition of complex-type N-glycans by Endoglycosidase S. Nat. Commun. 2018, 9, 1874.
- Aghababa, H.; Ting, Y.T.; Pilapitiya, D.; Loh, J.M.S.; Young, P.G.; Proft, T. Complement evasion factor (CEF), a novel immune evasion factor of Streptococcus pyogenes. Virulence 2022, 13, 225–240.
- Åkesson, P.; Sjöholm, A.G.; Björck, L. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J. Biol. Chem. 1996, 271, 1081–1088.
- Terao, Y.; Mori, Y.; Yamaguchi, M.; Shimizu, Y.; Ooe, K.; Hamada, S.; Kawabata, S. Group A streptococcal cysteine protease degrades C3 (C3b) and contributes to evasion of innate immunity. J. Biol. Chem. 2008, 283, 6253–6260.
- Kuo, C.-F.; Lin, Y.-S.; Chuang, W.-J.; Wu, J.-J.; Tsao, N. Degradation of complement 3 by streptococcal pyrogenic exotoxin B inhibits complement activation and neutrophil opsonophagocytosis. Infect. Immun. 2008, 76, 1163–1169.
- Tsao, N.; Tsai, W.-H.; Lin, Y.-S.; Chuang, W.-J.; Wang, C.-H.; Kuo, C.-F. Streptococcal pyrogenic exotoxin B cleaves properdin and inhibits complement-mediated opsonophagocytosis. Biochem. Biophys. Res. Commun. 2006, 339, 779–784.
- Cleary, P.P.; Prahbu, U.; Dale, J.B.; Wexler, D.E.; Handley, J. Streptococcal C5a peptidase is a highly specific endopeptidase. Infect. Immun. 1992, 60, 5219–5223.
- Wexler, D.E.; Cleary, P.P. Purification and characteristics of the streptococcal chemotactic factor inactivator. Infect. Immun. 1985, 50, 757–764.
- DeMaster, E.; Schnitzler, N.; Cheng, Q.; Cleary, P. M+ group a streptococci are phagocytized and killed in whole blood by C5a-activated polymorphonuclear leukocytes. Infect. Immun. 2002, 70, 350–359.
- Berge, A.; Björck, L. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J. Biol. Chem. 1995, 270, 9862–9867.
- Yang, D.; Chen, Q.; Hoover, D.; Staley, P.; Tucker, K.; Lubkowski, J.; Oppenheim, J. Many chemokines including CCL20/MIP-3α display antimicrobial activity. J. Leukoc. Biol. 2003, 74, 448–455.
- Kapur, V.; Majesky, M.W.; Li, L.L.; Black, R.A.; Musser, J.M. Cleavage of interleukin 1β (IL-1β) precursor to produce active IL-1β by a conserved extracellular cysteine protease from Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 1993, 90, 7676–7680.
- Herwald, H.; Collin, M.; Müller-Esterl, W.; Björck, L. Streptococcal cysteine proteinase releases kinins: A novel virulence mechanism. J. Exp. Med. 1996, 184, 665–673.
- Watanabe, Y.; Todome, Y.; Ohkuni, H.; Sakurada, S.; Ishikawa, T.; Yutsudo, T.; Fischetti, V.A.; Zabriskie, J.B. Cysteine protease activity and histamine release from the human mast cell line HMC-1 stimulated by recombinant streptococcal pyrogenic exotoxin B/streptococcal cysteine protease. Infect. Immun. 2002, 70, 3944–3947.
- Hidalgo-Grass, C.; Dan-Goor, M.; Maly, A.; Eran, Y.; Kwinn, L.A.; Nizet, V.; Ravins, M.; Jaffe, J.; Peyser, A.; Moses, A.E.; et al. Effect of a bacterial pheromone peptide on host chemokine degradation in group A streptococcal necrotising soft-tissue infections. Lancet 2004, 363, 696–703.
- Turner, C.E.; Kurupati, P.; Jones, M.D.; Edwards, R.J.; Sriskandan, S. Emerging role of the interleukin-8 cleaving enzyme SpyCEP in clinical Streptococcus pyogenes infection. J. Infect. Dis. 2009, 200, 555–563.
- Zingaretti, C.; Falugi, F.; Nardi-Dei, V.; Pietrocola, G.; Mariani, M.; Liberatori, S.; Gallotta, M.; Tontini, M.; Tani, C.; Speziale, P.; et al. Streptococcus pyogenes SpyCEP: A chemokine-inactivating protease with unique structural and biochemical features. FASEB J. 2010, 24, 2839–2848.
- Chiappini, N.; Seubert, A.; Telford, J.L.; Grandi, G.; Serruto, D.; Margarit, I.; Janulczyk, R. Streptococcus pyogenes SpyCEP influences host-pathogen interactions during infection in a murine air pouch model. PLoS ONE 2012, 7, e40411.
- Lawrenson, R.A.; Sriskandan, S. Cell Envelope Proteinase A (Streptococcus). Handbook of Proteolytic Enzymes; Elsevier: Amsterdam, The Netherlands, 2013; pp. 3195–3202.
- Edwards, R.J.; Taylor, G.W.; Ferguson, M.; Murray, S.; Rendell, N.; Wrigley, A.; Bai, Z.; Boyle, J.; Finney, S.J.; Jones, A.; et al. Specific C-terminal cleavage and inactivation of interleukin-8 by invasive disease isolates of Streptococcus pyogenes. J. Infect. Dis. 2005, 192, 783–790.
- Sumby, P.; Zhang, S.; Whitney, A.R.; Falugi, F.; Grandi, G.; Graviss, E.A.; Deleo, F.R.; Musser, J.M. A chemokine-degrading extracellular protease made by group A Streptococcus alters pathogenesis by enhancing evasion of the innate immune response. Infect. Immun. 2008, 76, 978–985.
- Zinkernagel, A.S.; Timmer, A.M.; Pence, M.A.; Locke, J.B.; Buchanan, J.T.; Turner, C.E.; Mishalian, I.; Sriskandan, S.; Hanski, E.; Nizet, V. The IL-8 protease SpyCEP/ScpC of group A Streptococcus promotes resistance to neutrophil killing. Cell Host Microbe 2008, 4, 170–178.
- Soderholm, A.T.; Barnett, T.C.; Korn, O.; Rivera-Hernandez, T.; Seymour, L.M.; Schulz, B.L.; Nizet, V.; Wells, C.A.; Sweet, M.J.; Walker, M.J. Group A Streptococcus M1T1 intracellular infection of primary tonsil epithelial cells dampens levels of secreted IL-8 through the action of SpyCEP. Front. Cell. Infect. Microbiol. 2018, 8, 160.
- Biswas, D.; Ambalavanan, P.; Ravins, M.; Anand, A.; Sharma, A.; Lim, K.X.Z.; Tan, R.Y.M.; Lim, H.Y.; Sol, A.; Bachrach, G.; et al. LL-37-mediated activation of host receptors is critical for defense against group A streptococcal infection. Cell Rep. 2021, 34, 108766.
- Jobichen, C.; Tan, Y.C.; Prabhakar, M.T.; Nayak, D.; Biswas, D.; Pannu, N.S.; Hanski, E.; Sivaraman, J. Structure of ScpC, a virulence protease from Streptococcus pyogenes, reveals the functional domains and maturation mechanism. Biochem. J. 2018, 475, 2847–2860.
- Kagawa, T.F.; O’Connell, M.R.; Mouat, P.; Paoli, M.; O’Toole, P.W.; Cooney, J.C. Model for substrate interactions in C5a peptidase from Streptococcus pyogenes: A 1.9 Å crystal structure of the active form of ScpA. J. Mol. Biol. 2009, 386, 754–772.
- Aziz, R.K.; Ismail, S.A.; Park, H.-W.; Kotb, M. Post-proteomic identification of a novel phage-encoded streptodornase, Sda1, in invasive M1T1 Streptococcus pyogenes. Mol. Microbiol. 2004, 54, 184–197.
- Aziz, R.K.; Edwards, R.A.; Taylor, W.W.; Low, D.E.; McGeer, A.; Kotb, M. Mosaic prophages with horizontally acquired genes account for the emergence and diversification of the globally disseminated M1T1 clone of Streptococcus pyogenes. J. Bacteriol. 2005, 187, 3311–3318.
- Buchanan, J.T.; Simpson, A.J.; Aziz, R.K.; Liu, G.Y.; Kristian, S.A.; Kotb, M.; Feramisco, J.; Nizet, V. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr. Biol. 2006, 16, 396–400.
- Uchiyama, S.; Andreoni, F.; Schuepbach, R.A.; Nizet, V.; Zinkernagel, A.S. DNase Sda1 allows invasive M1T1 Group A Streptococcus to prevent TLR9-dependent recognition. PLoS Pathog. 2012, 8, e1002736.
- Moon, A.F.; Krahn, J.M.; Lu, X.; Cuneo, M.J.; Pedersen, L.C. Structural characterization of the virulence factor Sda1 nuclease from Streptococcus pyogenes. Nucleic Acids Res. 2016, 44, 3946–3957.
- Chang, A.; Khemlani, A.; Kang, H.; Proft, T. Functional analysis of Streptococcus pyogenes nuclease A (SpnA), a novel group A streptococcal virulence factor. Mol. Microbiol. 2011, 79, 1629–1642.
- Hasegawa, T.; Minami, M.; Okamoto, A.; Tatsuno, I.; Isaka, M.; Ohta, M. Characterization of a virulence-associated and cell-wall-located DNase of Streptococcus pyogenes. Microbiology 2010, 156 Pt 1, 184–190.
- Zheng, L.; Khemlani, A.; Lorenz, N.; Loh, J.M.S.; Langley, R.J.; Proft, T. Streptococcal 5′-Nucleotidase A (S5nA), a novel Streptococcus pyogenes virulence factor that facilitates immune evasion. J. Biol. Chem. 2015, 290, 31126–31137.
- Firon, A.; Dinis, M.; Raynal, B.; Poyart, C.; Trieu-Cuot, P.; Kaminski, P.A. Extracellular nucleotide catabolism by the Group B Streptococcus ectonucleotidase NudP increases bacterial survival in blood. J. Biol. Chem. 2014, 289, 5479–5489.
- Thammavongsa, V.; Schneewind, O.; Missiakas, D.M. Enzymatic properties of Staphylococcus aureus adenosine synthase (AdsA). BMC Biochem. 2011, 12, 56.
- Fan, J.; Zhang, Y.; Chuang-Smith, O.N.; Frank, K.L.; Guenther, B.D.; Kern, M.; Schlievert, P.M.; Herzberg, M.C. Ecto-5′-nucleotidase: A candidate virulence factor in Streptococcus sanguinis experimental endocarditis. PLoS ONE 2012, 7, e38059.
- Dangel, M.-L.; Dettmann, J.-C.; Haßelbarth, S.; Krogull, M.; Schakat, M.; Kreikemeyer, B.; Fiedler, T. The 5′-nucleotidase S5nA is dispensable for evasion of phagocytosis and biofilm formation in Streptococcus pyogenes. PLoS ONE 2019, 14, e0211074.
- Madden, J.C.; Ruiz, N.; Caparon, M. Cytolysin-mediated translocation (CMT): A functional equivalent of type III secretion in gram-positive bacteria. Cell 2001, 104, 143–152.
- Meehl, M.A.; Pinkner, J.S.; Anderson, P.J.; Hultgren, S.J.; Caparon, M.G. A novel endogenous inhibitor of the secreted streptococcal NAD-glycohydrolase. PLoS Pathog. 2005, 1, e35.
- Yoon, J.Y.; An, D.R.; Yoon, H.J.; Kim, H.S.; Lee, S.J.; Im, H.N.; Jang, J.Y.; Suh, S.W. High-resolution crystal structure of Streptococcus pyogenes β-NAD+ glycohydrolase in complex with its endogenous inhibitor IFS reveals a highly water-rich interface. J. Synchrotron. Radiat. 2013, 20 Pt 6, 962–967.
- Velarde, J.J.; Piai, A.; Lichtenstein, I.J.; Lynskey, N.N.; Chou, J.J.; Wessels, M.R. Structure of the Streptococcus pyogenes NAD+ glycohydrolase translocation domain and its essential role in toxin binding to oropharyngeal keratinocytes. J. Bacteriol. 2022, 204, e0036621.
- Fahmi, T.; Faozia, S.; Port, G.; Cho, K.H. The second messenger c-di-AMP regulates diverse cellular pathways involved in stress response, biofilm formation, cell wall Homeostasis, SpeB expression and virulence in Streptococcus pyogenes. Infect. Immun. 2019, 87, e00147-19.
- Movert, E.; Bolarin, J.S.; Valfridsson, C.; Velarde, J.; Skrede, S.; Nekludov, M.; Hyldegaard, O.; Arnell, P.; Svensson, M.; Norrby-Teglund, A.; et al. Interplay between human STING genotype and bacterial NADase activity regulates inter-individual disease variability. Nat. Commun. 2023, 14, 4008.
- Broz, P.; Pelegrín, P.; Shao, F. The gasdermins, a protein family executing cell death and inflammation. Nat. Rev. Immunol. 2020, 20, 143–157.
- LaRock, D.L.; Johnson, A.F.; Wilde, S.; Sands, J.S.; Monteiro, M.P.; LaRock, C.N. Group A Streptococcus induces GSDMA-dependent pyroptosis in keratinocytes. Nature 2022, 605, 527–531.
- Barnett, T.C.; Liebl, D.; Seymour, L.M.; Gillen, C.M.; Lim, J.Y.; Larock, C.N.; Davies, M.R.; Schulz, B.L.; Nizet, V.; Teasdale, R.D.; et al. The globally disseminated M1T1 clone of group A Streptococcus evades autophagy for intracellular replication. Cell Host Microbe 2013, 14, 675–682.
- Kanaoka, M.; Fukita, Y.; Taya, K.; Kawanaka, C.; Negoro, T.; Agui, H. Antitumor activity of streptococcal acid glycoprotein produced by Streptococcus pyogenes Su. Jpn. J. Cancer Res. 1987, 78, 1409–1414.
- Degnan, B.A.; Palmer, J.M.; Robson, T.; Jones, C.E.; Fischer, M.; Glanville, M.; Mellor, G.D.; Diamond, A.G.; Kehoe, M.A.; Goodacre, J.A. Inhibition of human peripheral blood mononuclear cell proliferation by Streptococcus pyogenes cell extract is associated with arginine deiminase activity. Infect. Immun. 1998, 66, 3050–3058.
- Starikova, E.A.; Mammedova, J.T.; Ozhiganova, A.; Leveshko, T.A.; Lebedeva, A.M.; Sokolov, A.V.; Isakov, D.V.; Karaseva, A.B.; Burova, L.A.; Kudryavtsev, I.V. Streptococcal arginine deiminase inhibits T lymphocyte differentiation in vitro. Microorganisms 2023, 11, 2585.
- Degnan, B.A.; Fontaine, M.C.; Doebereiner, A.H.; Lee, J.J.; Mastroeni, P.; Dougan, G.; Goodacre, J.A.; Kehoe, M.A. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect. Immun. 2000, 68, 2441–2448.
- Henningham, A.; Ericsson, D.J.; Langer, K.; Casey, L.W.; Jovcevski, B.; Chhatwal, G.S.; Aquilina, J.A.; Batzloff, M.R.; Kobe, B.; Walker, M.J. Structure-informed design of an enzymatically inactive vaccine component for group A Streptococcus. MBio 2013, 4, e00509-13.




