
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Emir Begagić | -- | 3610 | 2024-01-25 11:43:49 | | | |
| 2 | Fanny Huang | Meta information modification | 3610 | 2024-01-30 06:41:40 | | |
Video Upload Options
The complexity of CRISPR-Cas9 applications in GBM research is highlighted, providing unique insights into apoptosis, cell proliferation, and immune responses within the tumor microenvironment. The studies challenge conventional perspectives on specific genes, emphasizing the potential therapeutic implications of manipulating key molecular players in cell cycle dynamics. Exploring CRISPR/Cas9 gene therapy in GBMs yields significant insights into the regulation of cellular processes, spanning cell interphase, renewal, and migration. Researchers, by precisely targeting specific genes, uncover the molecular orchestration governing cell proliferation, growth, and differentiation during critical phases of the cell cycle. The findings underscore the potential of CRISPR/Cas9 technology in unraveling the complex dynamics of the GBM microenvironment, offering promising avenues for targeted therapies to curb GBM growth.
1. Introduction
2. CRISPR/Cas9-Mediated GBM Therapy
2.1. Targeting Specific Genetic Mutations in GBM
2.1.1. Cell Cycle Regulation
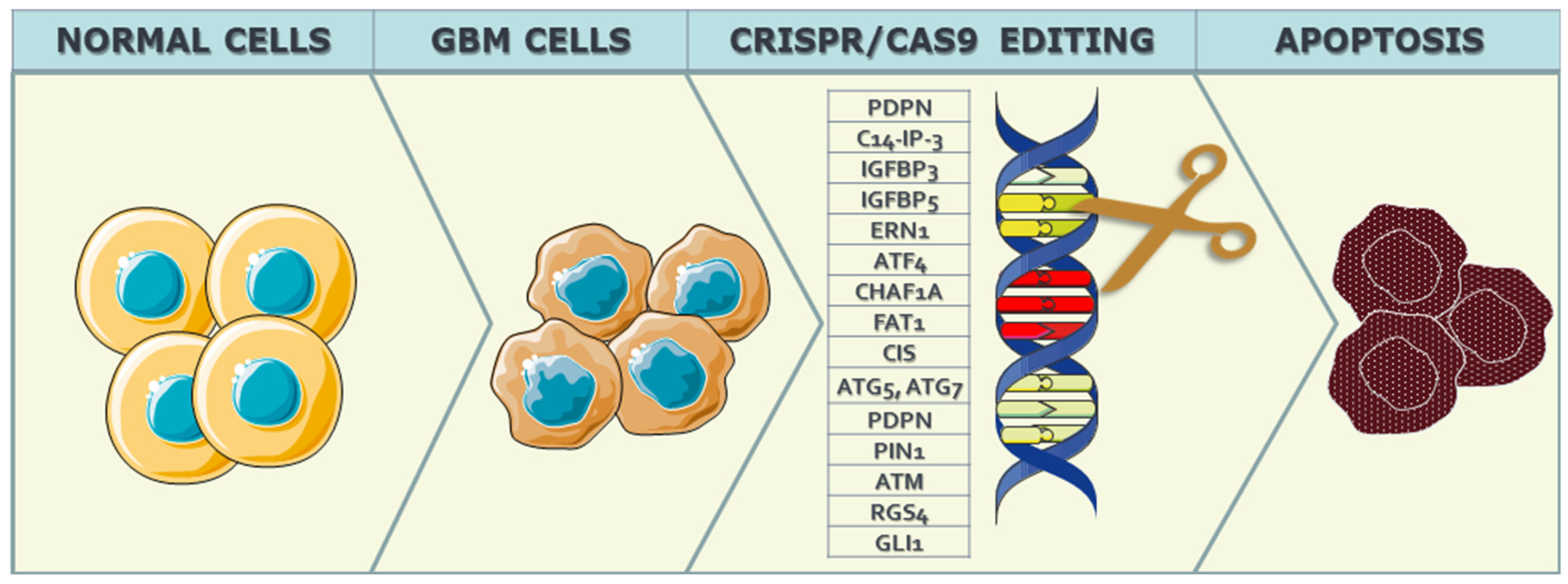
2.1.2. Cell-Interphase-Related Targets
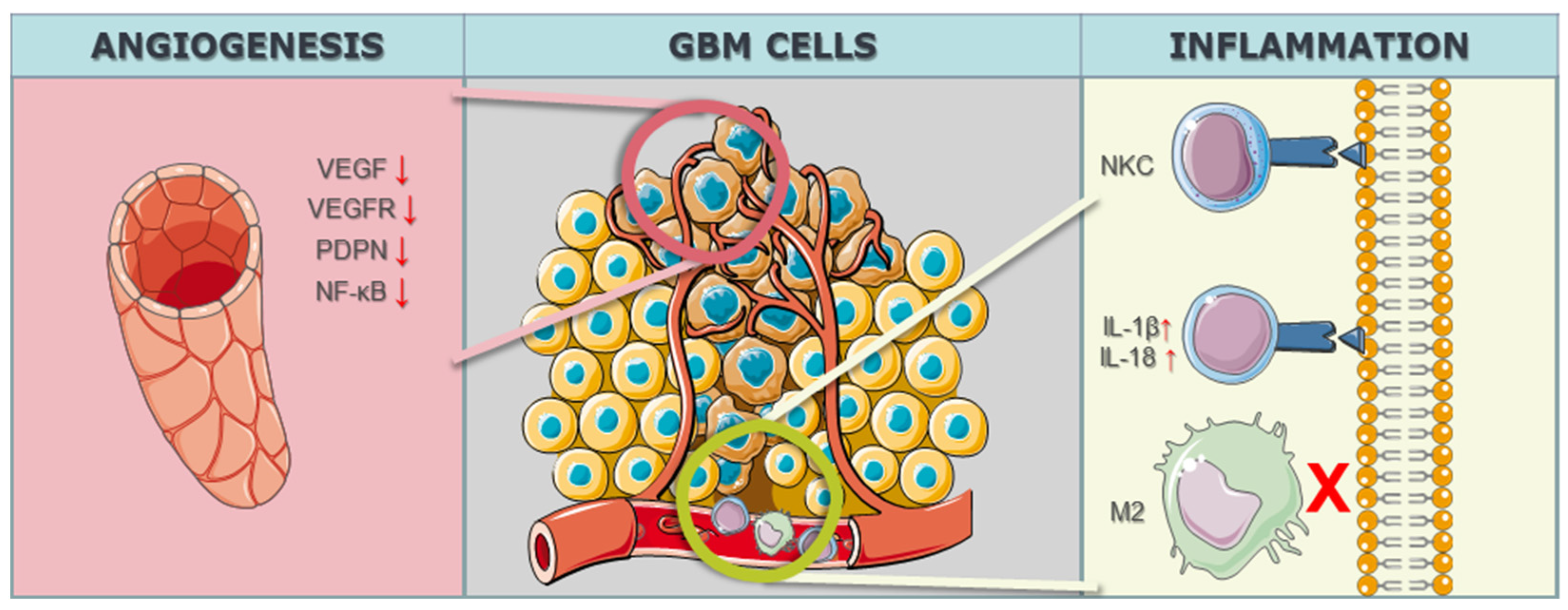
2.1.3. Microenvironmental CRISPR/Cas9 Targets in GBM Cells
2.2. Contribution of CRISPR/Cas9 Technology in Alleviating Therapy Resistance of GBM
References
- Begagić, E.; Pugonja, R.; Bečulić, H.; Čeliković, A.; Tandir Lihić, L.; Kadić Vukas, S.; Čejvan, L.; Skomorac, R.; Selimović, E.; Jaganjac, B.; et al. Molecular Targeted Therapies in Glioblastoma Multiforme: A Systematic Overview of Global Trends and Findings. Brain Sci. 2023, 13, 1602.
- Stoyanov, G.S.; Lyutfi, E.; Georgieva, R.; Georgiev, R.; Dzhenkov, D.L.; Petkova, L.; Ivanov, B.D.; Kaprelyan, A.; Ghenev, P. Reclassification of Glioblastoma Multiforme According to the 2021 World Health Organization Classification of Central Nervous System Tumors: A Single Institution Report and Practical Significance. Cureus 2022, 14, e21822.
- Jain, K.K. A Critical Overview of Targeted Therapies for Glioblastoma. Front. Oncol. 2018, 8, 419.
- Tatebayashi, K.; Nakayama, N.; Sakamoto, D.; Iida, T.; Ono, S.; Matsuda, I.; Enomoto, Y.; Tanaka, M.; Fujita, M.; Hirota, S.; et al. Clinical Significance of Early Venous Filling Detected via Preoperative Angiography in Glioblastoma. Cancers 2023, 15, 3800.
- Angom, R.S.; Nakka, N.M.R.; Bhattacharya, S. Advances in Glioblastoma Therapy: An Update on Current Approaches. Brain Sci. 2023, 13, 1536.
- Agosti, E.; Zeppieri, M.; De Maria, L.; Tedeschi, C.; Fontanella, M.M.; Panciani, P.P.; Ius, T. Glioblastoma Immunotherapy: A Systematic Review of the Present Strategies and Prospects for Advancements. Int. J. Mol. Sci. 2023, 24, 15037.
- Riemenschneider, M.J.; Jeuken, J.W.; Wesseling, P.; Reifenberger, G. Molecular diagnostics of gliomas: State of the art. Acta Neuropathol. 2010, 120, 567–584.
- Isachesku, E.; Braicu, C.; Pirlog, R.; Kocijancic, A.; Busuioc, C.; Pruteanu, L.-L.; Pandey, D.P.; Berindan-Neagoe, I. The Role of Non-Coding RNAs in Epigenetic Dysregulation in Glioblastoma Development. Int. J. Mol. Sci. 2023, 24, 16320.
- Senhaji, N.; Squalli Houssaini, A.; Lamrabet, S.; Louati, S.; Bennis, S. Molecular and Circulating Biomarkers in Patients with Glioblastoma. Int. J. Mol. Sci. 2022, 23, 7474.
- Khlidj, Y. What did CRISPR-Cas9 accomplish in its first 10 years? Biochem. Med. 2023, 33, 030601.
- Peixoto, J.; Príncipe, C.; Pestana, A.; Osório, H.; Pinto, M.T.; Prazeres, H.; Soares, P.; Lima, R.T. Using a Dual CRISPR/Cas9 Approach to Gain Insight into the Role of LRP1B in Glioblastoma. Int. J. Mol. Sci. 2023, 24, 11285.
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477.
- Gallo, K.; Srinageshwar, B.; Ward, A.; Diola, C.; Dunbar, G.; Rossignol, J.; Bakke, J. Inducible Knockout of 14-3-3β Attenuates Proliferation and Spheroid Formation in a Human Glioblastoma Cell Line U87MG. Brain Sci. 2023, 13, 868.
- Motoche-Monar, C.; Ordoñez, J.E.; Chang, O.; Gonzales-Zubiate, F.A. gRNA Design: How Its Evolution Impacted on CRISPR/Cas9 Systems Refinement. Biomolecules 2023, 13, 1698.
- Ding, S.; Liu, J.; Han, X.; Tang, M. CRISPR/Cas9-Mediated Genome Editing in Cancer Therapy. Int. J. Mol. Sci. 2023, 24, 16325.
- Begagić, E.; Pugonja, R.; Bečulić, H.; Selimović, E.; Skomorac, R.; Saß, B.; Pojskić, M. The new era of spinal surgery: Exploring the utilization of exoscopes as a viable alternative to operative microscopes—A systematic review and meta-analysis. World Neurosurg. 2023, in press.
- Begagić, E.; Bečulić, H.; Skomorac, R.; Pojskić, M. Accessible Spinal Surgery: Transformation Through the Implementation of Exoscopes As Substitutes for Conventional Microsurgery in Low- and Middle-Income Settings. Cureus 2023, 15, e45350.
- Kang, X.; Wang, Y.; Liu, P.; Huang, B.; Zhou, B.; Lu, S.; Geng, W.; Tang, H. Progresses, Challenges, and Prospects of CRISPR/Cas9 Gene-Editing in Glioma Studies. Cancers 2023, 15, 396.
- Qazi, M.A.; Vora, P.; Venugopal, C.; Sidhu, S.S.; Moffat, J.; Swanton, C.; Singh, S.K. Intratumoral heterogeneity: Pathways to treatment resistance and relapse in human glioblastoma. Ann. Oncol. 2017, 28, 1448–1456.
- Lake, J.A.; Donson, A.M.; Prince, E.; Davies, K.D.; Nellan, A.; Green, A.L.; Mulcahy Levy, J.; Dorris, K.; Vibhakar, R.; Hankinson, T.C.; et al. Targeted fusion analysis can aid in the classification and treatment of pediatric glioma, ependymoma, and glioneuronal tumors. Pediatr. Blood Cancer 2020, 67, e28028.
- Wang, X.; Wang, X.; Li, J.; Liang, J.; Ren, X.; Yun, D.; Liu, J.; Fan, J.; Zhang, Y.; Zhang, J.; et al. PDPN contributes to constructing immunosuppressive microenvironment in IDH wildtype glioma. Cancer Gene Therapy 2023, 30, 345–357.
- Rodvold, J.J.; Xian, S.; Nussbacher, J.; Tsui, B.; Cameron Waller, T.; Searles, S.C.; Lew, A.; Jiang, P.; Babic, I.; Nomura, N.; et al. IRE1α and IGF signaling predict resistance to an endoplasmic reticulum stress-inducing drug in glioblastoma cells. Sci. Rep. 2020, 10, 8348.
- Vu, H.T.; Kobayashi, M.; Hegazy, A.M.; Tadokoro, Y.; Ueno, M.; Kasahara, A.; Takase, Y.; Nomura, N.; Peng, H.; Ito, C.; et al. Autophagy inhibition synergizes with calcium mobilization to achieve efficient therapy of malignant gliomas. Cancer Sci. 2018, 109, 2497–2508.
- Peng, H.; Du, B.; Jiang, H.; Gao, J. Over-expression of CHAF1A promotes cell proliferation and apoptosis resistance in glioblastoma cells via AKT/FOXO3a/Bim pathway. Biochem. Biophys. Res. Commun. 2016, 469, 1111–1116.
- Kranz, D.; Boutros, M. A synthetic lethal screen identifies FAT1 as an antagonist of caspase-8 in extrinsic apoptosis. Embo J. 2014, 33, 181–197.
- Nakazawa, T.; Morimoto, T.; Maeoka, R.; Matsuda, R.; Nakamura, M.; Nishimura, F.; Ouji, N.; Yamada, S.; Nakagawa, I.; Park, Y.S.; et al. CIS deletion by CRISPR/Cas9 enhances human primary natural killer cell functions against allogeneic glioblastoma. J. Exp. Clin. Cancer Res. 2023, 42, 205.
- Zielke, S.; Meyer, N.; Mari, M.; Abou-El-Ardat, K.; Reggiori, F.; van Wijk, S.J.L.; Kögel, D.; Fulda, S. Loperamide, pimozide, and STF-62247 trigger autophagy-dependent cell death in glioblastoma cells. Cell. Death Dis. 2018, 9, 994.
- Ali, R.; Alabdullah, M.; Miligy, I.; Normatova, M.; Babaei-Jadidi, R.; Nateri, A.S.; Rakha, E.A.; Madhusudan, S. ATM Regulated PTEN Degradation Is XIAP E3 Ubiquitin Ligase Mediated in p85α Deficient Cancer Cells and Influence Platinum Sensitivity. Cells 2019, 8, 1271.
- Ranjan, A.; Srivastava, S.K. Penfluridol suppresses glioblastoma tumor growth by Akt-mediated inhibition of GLI1. Oncotarget 2017, 8, 32960–32976.
- Esemen, Y.; Awan, M.; Parwez, R.; Baig, A.; Rahman, S.; Masala, I.; Franchini, S.; Giakoumettis, D. Molecular Pathogenesis of Glioblastoma in Adults and Future Perspectives: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 2607.
- Fierro, J., Jr.; DiPasquale, J.; Perez, J.; Chin, B.; Chokpapone, Y.; Tran, A.M.; Holden, A.; Factoriza, C.; Sivagnanakumar, N.; Aguilar, R.; et al. Dual-sgRNA CRISPR/Cas9 knockout of PD-L1 in human U87 glioblastoma tumor cells inhibits proliferation, invasion, and tumor-associated macrophage polarization. Sci. Rep. 2022, 12, 2417.
- Lumibao, J.C.; Haak, P.L.; Kolossov, V.L.; Chen, J.E.; Stutchman, J.; Ruiz, A.; Sivaguru, M.; Sarkaria, J.N.; Harley, B.A.C.; Steelman, A.J.; et al. CHCHD2 mediates glioblastoma cell proliferation, mitochondrial metabolism, hypoxia-induced invasion and therapeutic resistance. Int. J. Oncol. 2023, 63, 117.
- Toledano, S.; Sabag, A.D.; Ilan, N.; Liburkin-Dan, T.; Kessler, O.; Neufeld, G. Plexin-A2 enables the proliferation and the development of tumors from glioblastoma derived cells. Cell Death Dis. 2023, 14, 41.
- Harutyunyan, A.S.; Krug, B.; Chen, H.; Papillon-Cavanagh, S.; Zeinieh, M.; De Jay, N.; Deshmukh, S.; Chen, C.C.L.; Belle, J.; Mikael, L.G.; et al. H3K27M induces defective chromatin spread of PRC2-mediated repressive H3K27me2/me3 and is essential for glioma tumorigenesis. Nat. Commun. 2019, 10, 1262.
- Guda, M.R.; Velpula, K.K.; Asuthkar, S.; Cain, C.P.; Tsung, A.J. Targeting RGS4 Ablates Glioblastoma Proliferation. Int. J. Mol. Sci. 2020, 21, 3300.
- Zhang, F.; Liu, R.; Liu, C.; Zhang, H.; Lu, Y. Nanos3, a cancer-germline gene, promotes cell proliferation, migration, chemoresistance, and invasion of human glioblastoma. Cancer Cell. Int. 2020, 20, 197.
- Godoy, P.R.D.V.; Pour Khavari, A.; Rizzo, M.; Sakamoto-Hojo, E.T.; Haghdoost, S. Targeting NRF2, Regulator of Antioxidant System, to Sensitize Glioblastoma Neurosphere Cells to Radiation-Induced Oxidative Stress. Oxidative Med. Cell. Longev. 2020, 2020, 2534643.
- Zhang, F.; Liu, R.; Zhang, H.; Liu, C.; Liu, C.; Lu, Y. Suppressing Dazl modulates tumorigenicity and stemness in human glioblastoma cells. BMC Cancer 2020, 20, 673.
- Liu, J.; Sareddy, G.R.; Zhou, M.; Viswanadhapalli, S.; Li, X.; Lai, Z.; Tekmal, R.R.; Brenner, A.; Vadlamudi, R.K. Differential Effects of Estrogen Receptor β Isoforms on Glioblastoma Progression. Cancer Res. 2018, 78, 3176–3189.
- Bulstrode, H.; Johnstone, E.; Marques-Torrejon, M.A.; Ferguson, K.M.; Bressan, R.B.; Blin, C.; Grant, V.; Gogolok, S.; Gangoso, E.; Gagrica, S.; et al. Elevated FOXG1 and SOX2 in glioblastoma enforces neural stem cell identity through transcriptional control of cell cycle and epigenetic regulators. Genes. Dev. 2017, 31, 757–773.
- Saenz-Antoñanzas, A.; Moncho-Amor, V.; Auzmendi-Iriarte, J.; Elua-Pinin, A.; Rizzoti, K.; Lovell-Badge, R.; Matheu, A. CRISPR/Cas9 Deletion of SOX2 Regulatory Region 2 (SRR2) Decreases SOX2 Malignant Activity in Glioblastoma. Cancers 2021, 13, 1574.
- Ogawa, J.; Pao, G.M.; Shokhirev, M.N.; Verma, I.M. Glioblastoma Model Using Human Cerebral Organoids. Cell. Rep. 2018, 23, 1220–1229.
- Smolkin, T.; Nir-Zvi, I.; Duvshani, N.; Mumblat, Y.; Kessler, O.; Neufeld, G. Complexes of plexin-A4 and plexin-D1 convey semaphorin-3C signals to induce cytoskeletal collapse in the absence of neuropilins. J. Cell. Sci. 2018, 131, jcs208298.
- Prolo, L.M.; Li, A.; Owen, S.F.; Parker, J.J.; Foshay, K.; Nitta, R.T.; Morgens, D.W.; Bolin, S.; Wilson, C.M.; Vega, L.J.; et al. Targeted genomic CRISPR-Cas9 screen identifies MAP4K4 as essential for glioblastoma invasion. Sci. Rep. 2019, 9, 14020.
- Wang, Y.; Yang, C.H.; Schultz, A.P.; Sims, M.M.; Miller, D.D.; Pfeffer, L.M. Brahma-Related Gene-1 (BRG1) promotes the malignant phenotype of glioblastoma cells. J. Cell. Mol. Med. 2021, 25, 2956–2966.
- Shao, W.; Azam, Z.; Guo, J.; To, S.S.T. Oncogenic potential of PIK3CD in glioblastoma is exerted through cytoskeletal proteins PAK3 and PLEK2. Lab. Investig. 2022, 102, 1314–1322.
- Chen, L.; Fang, W.; Chen, W.; Wei, Y.; Ding, J.; Li, J.; Lin, J.; Wu, Q. Deciphering the molecular mechanism of the THBS1 gene in the TNF signaling axis in glioma stem cells. Cell. Signal. 2023, 106, 110656.
- Ezgi, O.-G.; Ezgi Yagmur, K.; Ali Cenk, A.; Ipek, B.; Ahmet, C.; Sheikh, N.; Martin, B.; Fidan, S.-P.; Tunc, M.; Can, A.; et al. Epigenetic-focused CRISPR/Cas9 screen identifies ASH2L as a regulator of glioblastoma cell survival. bioRxiv 2022. peer review.
- Nieland, L.; van Solinge, T.S.; Cheah, P.S.; Morsett, L.M.; El Khoury, J.; Rissman, J.I.; Kleinstiver, B.P.; Broekman, M.L.D.; Breakefield, X.O.; Abels, E.R. CRISPR-Cas knockout of miR21 reduces glioma growth. Mol. Ther. Oncolytics 2022, 25, 121–136.
- Uceda-Castro, R.; van Asperen, J.V.; Vennin, C.; Sluijs, J.A.; van Bodegraven, E.J.; Margarido, A.S.; Robe, P.A.J.; van Rheenen, J.; Hol, E.M. GFAP splice variants fine-tune glioma cell invasion and tumour dynamics by modulating migration persistence. Sci. Rep. 2022, 12, 424.
- Han, N.; Hu, G.; Shi, L.; Long, G.; Yang, L.; Xi, Q.; Guo, Q.; Wang, J.; Dong, Z.; Zhang, M. Notch1 ablation radiosensitizes glioblastoma cells. Oncotarget 2017, 8, 88059–88068.
- Eisemann, T.; Costa, B.; Harter, P.N.; Wick, W.; Mittelbronn, M.; Angel, P.; Peterziel, H. Podoplanin expression is a prognostic biomarker but may be dispensable for the malignancy of glioblastoma. Neuro-Oncol. 2019, 21, 326–336.
- Szymura, S.J.; Bernal, G.M.; Wu, L.; Zhang, Z.; Crawley, C.D.; Voce, D.J.; Campbell, P.A.; Ranoa, D.E.; Weichselbaum, R.R.; Yamini, B. DDX39B interacts with the pattern recognition receptor pathway to inhibit NF-κB and sensitize to alkylating chemotherapy. BMC Biol. 2020, 18, 32.
- Lu, F.I.; Wang, Y.T.; Wang, Y.S.; Wu, C.Y.; Li, C.C. Involvement of BIG1 and BIG2 in regulating VEGF expression and angiogenesis. FASEB J. 2019, 33, 9959–9973.
- Wei, J.; Marisetty, A.; Schrand, B.; Gabrusiewicz, K.; Hashimoto, Y.; Ott, M.; Grami, Z.; Kong, L.Y.; Ling, X.; Caruso, H.; et al. Osteopontin mediates glioblastoma-associated macrophage infiltration and is a potential therapeutic target. J. Clin. Investig. 2019, 129, 137–149.
- Chen, P.A.; Shrivastava, G.; Balcom, E.F.; McKenzie, B.A.; Fernandes, J.; Branton, W.G.; Wheatley, B.M.; Petruk, K.; van Landeghem, F.K.H.; Power, C. Absent in melanoma 2 regulates tumor cell proliferation in glioblastoma multiforme. J. Neurooncol. 2019, 144, 265–273.
- Wu, W.; Wu, Y.; Mayer, K.; von Rosenstiel, C.; Schecker, J.; Baur, S.; Würstle, S.; Liesche-Starnecker, F.; Gempt, J.; Schlegel, J. Lipid Peroxidation Plays an Important Role in Chemotherapeutic Effects of Temozolomide and the Development of Therapy Resistance in Human Glioblastoma. Transl. Oncol. 2020, 13, 100748.
- Han, X.; Abdallah, M.O.E.; Breuer, P.; Stahl, F.; Bakhit, Y.; Potthoff, A.L.; Pregler, B.E.F.; Schneider, M.; Waha, A.; Wüllner, U.; et al. Downregulation of MGMT expression by targeted editing of DNA methylation enhances temozolomide sensitivity in glioblastoma. Neoplasia 2023, 44, 100929.
- Tong, F.; Zhao, J.X.; Fang, Z.Y.; Cui, X.T.; Su, D.Y.; Liu, X.; Zhou, J.H.; Wang, G.X.; Qiu, Z.J.; Liu, S.Z.; et al. MUC1 promotes glioblastoma progression and TMZ resistance by stabilizing EGFRvIII. Pharmacol. Res. 2023, 187, 106606.
- Liu, X.; Cao, Z.; Wang, W.; Zou, C.; Wang, Y.; Pan, L.; Jia, B.; Zhang, K.; Zhang, W.; Li, W.; et al. Engineered Extracellular Vesicle-Delivered CRISPR/Cas9 for Radiotherapy Sensitization of Glioblastoma. ACS Nano 2023, 17, 16432–16447.
- Rocha, C.R.R.; Rocha, A.R.; Silva, M.M.; Gomes, L.R.; Latancia, M.T.; Andrade-Tomaz, M.; de Souza, I.; Monteiro, L.K.S.; Menck, C.F.M. Revealing Temozolomide Resistance Mechanisms via Genome-Wide CRISPR Libraries. Cells 2020, 9, 2573.
- Yin, J.; Wang, X.; Ge, X.; Ding, F.; Shi, Z.; Ge, Z.; Huang, G.; Zhao, N.; Chen, D.; Zhang, J.; et al. Hypoxanthine phosphoribosyl transferase 1 metabolizes temozolomide to activate AMPK for driving chemoresistance of glioblastomas. Nat. Commun. 2023, 14, 5913.





