Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Daniel Alejandro Caballero Cerbon | -- | 3617 | 2024-01-22 11:44:38 | | | |
| 2 | Lindsay Dong | Meta information modification | 3617 | 2024-01-23 01:54:43 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Caballero Cerbon, D.A.; Gebhard, L.; Dokuyucu, R.; Ertl, T.; Härtl, S.; Mazhar, A.; Weuster-Botz, D. The Bioproduction of L-Cysteine. Encyclopedia. Available online: https://encyclopedia.pub/entry/54186 (accessed on 28 February 2026).
Caballero Cerbon DA, Gebhard L, Dokuyucu R, Ertl T, Härtl S, Mazhar A, et al. The Bioproduction of L-Cysteine. Encyclopedia. Available at: https://encyclopedia.pub/entry/54186. Accessed February 28, 2026.
Caballero Cerbon, Daniel Alejandro, Leon Gebhard, Ruveyda Dokuyucu, Theresa Ertl, Sophia Härtl, Ayesha Mazhar, Dirk Weuster-Botz. "The Bioproduction of L-Cysteine" Encyclopedia, https://encyclopedia.pub/entry/54186 (accessed February 28, 2026).
Caballero Cerbon, D.A., Gebhard, L., Dokuyucu, R., Ertl, T., Härtl, S., Mazhar, A., & Weuster-Botz, D. (2024, January 22). The Bioproduction of L-Cysteine. In Encyclopedia. https://encyclopedia.pub/entry/54186
Caballero Cerbon, Daniel Alejandro, et al. "The Bioproduction of L-Cysteine." Encyclopedia. Web. 22 January, 2024.
Copy Citation
L-cysteine is a proteogenic amino acid with many applications in the pharmaceutical, food, animal feed, and cosmetic industries. Due to safety and environmental issues in extracting L-cysteine from animal hair and feathers, the fermentative production of L-cysteine offers an attractive alternative using renewable feedstocks.
L-cysteine
biosynthesis
metabolic engineering
fermentation
1. Introduction
L-cysteine is a nucleophilic amino acid with a sulfhydryl group, which enables the formation of disulfide bridges. It can be produced by most eukaryotic and prokaryotic organisms. This amino acid is semi-essential in humans, since it can be synthesized endogenously or directly absorbed from an L-cysteine-rich diet. In cells, cysteine can be produced via the reverse transsulfuration pathway from L-methionine [1] or from L-serine through acetylation and subsequent sulfhydration [2]. Figure 1 presents a step-by-step diagram for these pathways. The cells tightly control the intracellular L-cysteine concentration because already-low concentrations can be cytotoxic [3][4]. Intracellular L-cysteine plays a vital role as a precursor for protein synthesis, and for the production of glutathione, hydrogen sulfide, and taurine [5]. In humans and animals, L-cysteine is primarily found in the hair, nails, and skin, especially in collagen. Prokaryotic cells use L-cysteine mostly in proteins, in enzymes for the formation of iron–sulfur clusters, or as a source of sulfur for other macromolecules [6].
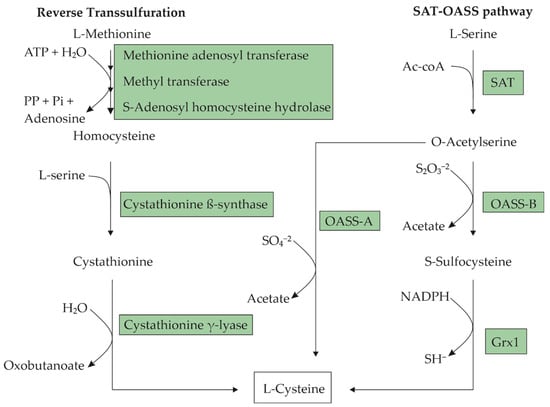
Figure 1. Metabolic pathways for L-cysteine biosynthesis in microorganisms. In the L-serine O-acetyltransferase-O-acetyl-L-serine (SAT-OASS) pathway, there are two branches for the transformation of O-acetyl-L-serine (OAS) into L-cysteine, depending on the sulfur source available to the organism. These biotransformations are catalyzed by two different OASSs: OASS-A in the case of sulfate as the S-source and OASS-B for thiosulfate. The transformation through OASS-B requires a subsequent reduction of S-sulfocysteine to L-cysteine catalyzed by glutathioredoxin (Grx1).
L-cysteine is produced for applications in the pharmaceutical, food, animal feed, and cosmetics industries, with an estimated yearly production of roughly 3000 tons [7][8]. In the pharmaceutical and food industries, L-cysteine is a precursor for the production of drugs, food supplements, and nutraceuticals [9]. In animal feed, L-cysteine is used as a sulfur supplement for wool production [10]. In the cosmetic industry, L-cysteine is applied as a substitute for thioglycolic acid in hair applications for breaking the disulfide bridges of keratin to achieve permanent hair waving [10].
Animal feathers and human hair are natural sources of L-cysteine, which is the main component of keratin. First, cystine is extracted with activated charcoal after the acidic hydrolysis of feathers and hair. After desorption from the activated charcoal matrix, the isolated and purified L-cystine is then transformed into L-cysteine via electrolytic reduction [11]. This is the most common production method applied so far, although the L-cysteine process yield is low, and unpleasant odors may occur. However, the most important disadvantage is the environmental and safety issues caused by the disposal of the organic waste, which contains hydrochloric acid [12].
An alternative approach is the biotechnological production of L-cysteine from renewable resources [13]. Since the 1980s, microorganisms of the genus Pseudomonas have been used for the asymmetric whole-cell biocatalysis of DL-2-amino-Δ2-thiazoline-4-carboxylic acid (DL-ATC) to generate L-cysteine (Figure 2).
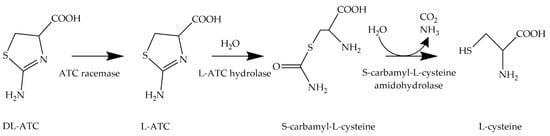
Figure 2. Transformation of DL-ATC to L-cysteine. In this enzymatic three-step process, DL-ATC is isomerized to L-ATC by ATC racemase. Then, L-ATC is hydrolyzed to S-carbamyl-L-cysteine (SCC) by L-ATC hydrolase. Finally, SCC is cleaved into L-cysteine, CO2, and ammonia by S-cabamyl-L-cysteine amidohydrolase.
Pseudomonas is a genus of gram-negative bacteria belonging to the family Pseudomonadaceae in the class of γ-proteobacteria. From the many bacteria, especially in the genus Pseudomonas, that have an activity to convert DL-ATC to L-cysteine, two biosynthetic pathways have been described [14][15][16]. As an intermediate, one pathway produces N-carbamoyl cysteine, and the other, S-carbamoyl cysteine [17]. However, in the Pseudomonas cells, L-cysteine is converted to pyruvate, hydrogen sulfide, and ammonia by L-cysteine desulfhydrase (CD). The L-cysteine desulfhydrase gene could not be knocked out to increase the production of L-cysteine during the bioconversion of DL-ATC by Pseudomonas sp., since the degradation of L-cysteine is required to supply the cells with carbon and nitrogen [18].
Through the understanding of the complex regulation system of the L-cysteine synthesis in bacteria, and with the advancements in synthetic biology, the direct fermentative L-cysteine production from glucose became feasible and has been exploited by global amino acid producers to supply an increasing portion of the global L-cysteine demand [19].
2. Metabolic Engineering Strategies
Microorganisms use two main pathways for the biosynthesis of L-cysteine. In enteric bacteria such as E. coli, the precursor of L-cysteine is L-serine [20]. L-cysteine is synthesized from L-serine in two steps, which involve the substitution of the β-hydroxyl group with a thiol group (Figure 1). The first step involves the acetylation of the β-hydroxyl of L-serine, resulting in an O-acetyl-L-serine (OAS) catalyzed by L-serine O-acetyltransferase (SAT), which is encoded by the cysE gene. The second step involves the α,β-elimination of acetate from OAS and the addition of H2S to give an L-cysteine catalyzed by O-acetyl-L-serine sulfhydrylase-A (OASS-A), which is encoded by the cysK gene. Corynebacterium glutamicum and Pantoea ananantis synthesize L-cysteine through the same pathway as E. coli [12]. Other microorganisms, such as Pseudomonas putida [21], Saccharomyces cerevisiae [22], and Lactococcus lactis [23] synthesize L-cysteine from L-methionine through the reverse transsulfuration pathway that consists in the cleavage of a methyl group from L-methionine to generate a homocysteine catalyzed by a multienzyme complex, and the homocysteine’s subsequent transformation to cystathionine by the enzyme cystathionine beta-synthase (CBS). L-cysteine can be produced from cystathionine through the enzyme cystathionine gamma-lyase (CSE) (Figure 1).
The choice of the starting strain for metabolic engineering has an impact on the potential enhancements achieved through the modification of metabolic steps [24]. This will be shown in an example with E. coli: It has been established that the transcription levels of key metabolic genes are the major modulators of different glucose utilization pathways in E. coli [25]. Also, the genetic difference in the catabolite repression protein (CRP) among the E. coli K-12 strains affects sulfur metabolism [26]. During the last 20 years, four E. coli K-12 strains (BW25113, JM109, W3110, and MG1655, respectively) have been utilized in L-cysteine production [27]. Therefore, the selection of a suitable chassis for L-cysteine production has been deemed the foundation for the successful implementation of metabolic engineering strategies [28]. To reach a more suitable chassis cell, the L-cysteine-producing plasmid pLH03, containing P-cysE (coding for serine acetyltransferase) and P-ydeD (a cysteine exporter), was transformed into the four aforementioned E. coli K-12 hosts. The results of shake flask experiments (minimal medium with thiosulfate as the sulfur source) showed that the best chassis for the plasmid pLH03 was BW25113, since it produced 345.8 mg L−1 of L-cysteine, followed by JM109 (190.3 mg L−1). By contrast, neither W3110 nor MG1655 were deemed suitable base strains for this plasmid, since they each produced less than 70 mg L−1 product. [28].
2.1. Biosynthesis of L-Cysteine
The acetylation of L-serine catalyzed by serine acetyltransferase (SAT) is the rate-limiting step of L-cysteine biosynthesis in many microorganisms. The activity of SAT is suppressed by L-cysteine via feedback inhibition (Figure 2). An approach to enhance the fermentative production of L-cysteine utilizes feedback inhibition-insensitive SATs. These SATs are obtained by the following means:
Nakamori et al. [33] obtained feedback inhibition-insensitive SATs by engineering SAT from E. coli through site-directed mutagenesis using PCR. In this study, the cysE gene coding for SAT was altered by replacing the methionine at position 256 with other amino acids. The transformed microorganisms, having the altered cysE gene, produced 50–300 mg L−1 of L-cysteine plus L-cystine after 72 h of cultivation in shake flasks containing a minimal medium at 30 °C. Furthermore, the transformation of the altered cysE gene in a mutant strain of E. coli, JM39-8, with decreased cysteine degradation abilities (10% CD activity) resulted in an increased production to a maximum of 790 mg L−1 of L-cysteine plus L-cystine in shake flasks under the same conditions as above.
Another approach to obtain feedback inhibition-insensitive SATs could be to use natural SATs from higher plants. In Arabidopsis thaliana, three cDNA clones encoding SAT isoforms (SAT-c, SAT-p, and SAT-m) have been isolated. Noji et al. [33] analyzed these SATs and reported that SAT-p and SAT-m were feedback inhibition-insensitive isozymes. However, the activity of SAT-c was feedback inhibited by a low concentration of L-cysteine.
C. glutamicum is a nonpathogenic and high G+C gram-positive bacterium industrially used for decades for the fermentative production of amino acids such as L-glutamic acid and L-lysine [34][35]. Joo et al. [36] investigated the effect of the overexpression of genes encoding SAT, OASS, and a transcriptional regulator in C. glutamicum. The results showed that L-cysteine production was enhanced by approximately three-fold, with the recombinant strain reaching a maximal L-cysteine concentration of approximately 60 mg L−1 in shake flasks at 30 °C after 15 h of cultivation.
One of the key enzymes in the L-serine biosynthesis pathway, 3-phosphoglycerate dehydrogenase (PGDH), coded by the gene serA, is feedback inhibited by L-serine [7][13]. The authors of [37] converted the 344th and 346th amino acids of E. coli’s PGDH to alanine, which created a feedback-insensitive serA.
2.2. Weakening the Degradation of L-Cysteine
It is well known that disrupting the degradation pathway and enhancing metabolic fluxes for the desired amino acids are effective approaches to improve their production [19][37][38][39]. The degradation of L-cysteine to pyruvate, ammonia, and sulfide is mainly catalyzed by L-cysteine desulfhydrase (CD). Wada et al. [40] purified and characterized CD from C. glutamicum. After comparing the partial amino acid sequence, it was identified that the enzyme CD is a C-S lyase with α,β-elimination activity, and it is encoded by the aecD gene [41]. They showed that the aecD gene product is involved in L-cysteine degradation, and the disruption of the aecD gene resulted in an increased production of L-cysteine (approximately 290 mg L−1 in shake flasks).
Awano et al. [42] investigated the enzyme having CD activity in E. coli using native-PAGE and CD activity staining. An analysis with gene-disrupted mutants showed that the tryptophanase (TNase) encoded by tnaA and cystathionine β-lyase (CBL) encoded by metC catalyze the degradation of L-cysteine in E. coli.
The transcriptional activator CysB, belonging to the LysR family, induces the transcription of genes in the sulfate pathway [43]. Kawano et al. [43] found a most plausible sequence for the CysB-binding motif upstream of the yciW gene. YciW encodes a soluble protein of 401 amino acids, although its function and structural information are still unknown. The CysB-binding motif is located at the −35 region of the predicted promotor, which is similar to that of cysJ coding for a sulfite reductase. The yciW-disrupted strain exhibited a much higher sensitivity to L-cysteine than the native strain. This indicates that yciW confers tolerance to cysteine in E. coli cells, and the authors suggest that yciW may convert L-cysteine to L-methionine or glutathione. Based on the deduced amino acid sequence, yciW is predicted to encode an oxidoreductase-like protein. YciW is also involved in the degradation of intracellular L-cysteine, leading to its detoxification. However, further investigation is needed at the protein level [43].
L-cysteine desulfidases are alternative L-cysteine-degrading enzymes that are less well characterized as compared to L-cysteine desulfhydrases. The activity of this enzyme was first observed in Methanocaldococcus jannaschii [44].
2.3. Regulation of L-Cysteine Transport
The bacterial production of metabolites depends on the equilibrium between secretion and reconsumption. High concentrations of L-cysteine have been reported to be toxic and inhibitory for microbial cells [3]. In E. coli, concentrations of 1 mM L-cysteine were enough to cause growth limitations [43]. Therefore, microorganisms have an exporter for L-cysteine within the L-cysteine regulon in order to avoid product-associated cytotoxicity.
Daßler et al. [45] identified the transporter protein YdeD in E. coli, which is reported to be one of the main facilitators involved in the export of L-cysteine. The corresponding gene encoding for the transporter protein involves an open reading frame coding for 299 amino acids. The substrate range of the YdeD protein is wide; therefore, it secretes other metabolites along with L-cysteine.
CydDC is an ATP-binding transporter reported to be involved in L-cysteine export from the cytoplasm to the periplasm in E. coli [46]. In a study, CydDC was compared to the gene product of ydeD and was found to more effectively remove high L-cysteine concentrations in intracellular compartments than ydeD. In shake flask experiments at 37 °C, E. coli mutants with deletions of cydDC accumulated 1.6 times higher L-cysteine concentrations than the wild type, while mutants with ydeD deletions accumulated 1.3 times higher L-cysteine concentrations. A double-deletion E. coli mutant of both cydDC and ydeD accumulated 2.2 times more L-cysteine than the wild type. [46]
yfiK was discovered to be a gene that augmented L-cysteine production when it was overexpressed in an E. coli production strain [47]. The gene product is an integral membrane protein with about six predicted transmembrane helices and belongs to the resistance to homoserine/threonine (RhtB) family of export proteins. YfiK overproduction from a plasmid leads to the drastic and parallel secretion of O-acetyl serine and L-cysteine into the medium, but only when the E. coli strain possesses an L-serine transacetylase that is feedback insensitive to L-cysteine. Externally provided, OAS obviated this requirement for L-cysteine secretion, both in the yfiK-carrying transformant and in the wild type. A ΔyfiK mutant did not show any phenotype, and this mutant exported OAS and L-cysteine when transformed with a plasmid carrying ydeD, a previously characterized, alternate OAS/L-cysteine exporter. Since a ydeD–yfiK double mutant showed the same pattern, it appears that YfiK and YdeD act independently. The necessity for the cell to regulate the size of the internal pool of OAS via the synthesis of exporter proteins could be connected to the fact that this compound inhibits growth when supplied externally. The overexpression of either ydeD or yfiK leads to alleviation of this inhibition parallel by an increased resistance to azaserine, which is an analog of OAS [47].
2.4. Limitations of the Metabolic Engineering Approaches
Liu et al. [13] developed an integrated approach, which used many of the previously presented metabolic engineering strategies in tandem. They overexpressed PSERT, PSL, and a feedback-insensitive version of PGDH, disrupted genes for the degradation of L-serine and L-cysteine, and overexpressed the L-cysteine exporter YdeD. Even though this strategy showed a cumulative improvement in the L-cysteine concentration, the final yield of L-cysteine on glucose was about 4.7% (w/w). This means that most of the fed substrate was not directly utilized for the L-cysteine production. Nonetheless, the authors indicate that this is the highest yield on glucose found in the literature at the time of its publication.
This exemplifies one of the major challenges of the metabolic engineering approaches. Most of the methods described above are applied directly on the L-cysteine synthesis pathway, which has been extensively characterized so that the bottlenecks are extensively identified. However, most metabolic pathways are thoroughly interconnected with regulation interactions that have not been yet characterized [48]. These interactions must then first be mapped out in order to identify further bottlenecks that can be tackled through rational metabolic engineering methods [48].
3. Process Engineering Aspects
3.1. Sulfur Assimilation Pathways in Bacteria
E. coli has two sulfur assimilation pathways for L-cysteine biosynthesis (Figure 2). One is the sulfate (SO42−) pathway, which consumes two molecules of ATP and four molecules of NADPH as a reducing power to produce L-cysteine from sulfate. The other is the thiosulfate (S2O32−) pathway, which spends only one molecule of NADPH from thiosulfate [49]. Thiosulfate is receiving attention as a sulfur source due to its advantageous effects on bacterial growth and L-cysteine production in comparison to sulfate [50].
Both sulfur assimilation pathways utilize OAS as a carbon skeleton to which they may incorporate sulfur. In the thiosulfate pathway, O-acetyl-L-serine sulfhydrylase B (CysM) catalyzes the conversion of OAS and thiosulfate into S-sulfocysteine, which is reduced into L-cysteine and sulfite by glutaredoxin (Grx1), and the glutaredoxin-like protein NrdH [49][51]. In comparison, sulfate is first reduced to sulfide in a four-step reaction catalyzed in subsequent order by sulfate adenylyltransferase (CysDN), adenylylsulfate kinase (CysC), 3′-phospho-adenylylsulfate reductase (CysH), and sulfite reductase (CysIJ). The subsequent incorporation of sulfide into OAS is enabled by O-acetyl-L-serine sulfhydrylase A (CysK) [52].
There is strong evidence suggesting that E. coli may possess an alternative pathway for assimilating thiosulfate, independent of the conventional pathway through the CysM enzyme [50]. This evidence comes from an experiment where ∆CysM E. coli cells were grown using thiosulfate as the only sulfur source. The ∆CysM cells showed a slower growth compared to the wild-type cells. However, when L-cystine was added to the growth media, the impaired growth was restored, indicating a deficiency in L-cysteine. The growth rate in the growth media without L-cystine was then increased by overexpressing thiosulfate sulfurtransferase (GlpE).
3.2. Glycerol as Carbon Source
Although glucose remains the predominant carbon source for L-cysteine biosynthesis, glycerol has gained increasing attention as an alternative carbon source due to its abundant availability as a by-product in biodiesel production [53]. Utilizing glycerol as a carbon source is crucial for reducing the costs of bioproduction and improving resource utilization. Moreover, the metabolic pathway from glycerol to L-cysteine is shorter and exhibits better carbon economy compared to glucose [54].
Significant progress has been made towards the bioproduction of L-cysteine from glycerol, as its essential precursor, L-serine, has already been produced effectively using glycerol [54]. The highest recorded concentration of L-cysteine from glycerol achieved so far is 313.4 mg L−1 with E. coli in shake flasks [54]. First, L-serine production was enhanced by making use of an L-serine biosensor based on the transcriptional regulator NCgl0581 for coupling L-serine biosynthesis to the growth rate of E. coli and using adaptive laboratory evolution (ALE).
3.3. Fed-Batch Processes in Stirred-Tank Bioreactors
Up to this point, most research presented had been carried out as simple batch processes in shake flasks, which allow for easier preparation and higher throughput than the scalable stirred-tank bioreactor. Investigations in fed-batch operated and fully controlled stirred-tank bioreactors using the observations provided by the studies in shake flasks and optimizing the reaction conditions may lead to an L-cysteine production scenario that is more in line with the expectations of the industry.
Liu et al. [13] utilized a rational approach which combined previously discussed metabolic engineering approaches to enhance the L-cysteine production of the E. coli strain JM109, including the overexpression of the enzymes in the L-serine biosynthetic pathway, feedback-insensitive phosphoglycerate dehydrogenase and serine acetyltransferase, the ydeD exporter, and the deletion of L-cysteine and serine deaminases. These modifications lead to a total L-cysteine production in shake flasks of 600 mg L−1. They then cultivated this modified strain in a 5 L stirred-tank reactor in a fed-batch operation, where a glucose solution was fed constantly into the reactor, but thiosulfate (the sulfur source) was only supplied at the beginning of the process. This led to an increase in the maximal achieved L-cysteine concentration to 5.1 g L−1.
3.4. Purification of L-Cysteine from the Fermentation Broth
L-cysteine, due to its high solubility in water and the vulnerability of its thiol group to oxidation, presents challenges in its purification for industrial applications. Several approaches have been proposed to address this issue [55].
Most approaches follow a two-stage process, which involves converting L-cysteine into L-cystine by treating the clarified solution with an oxidizing agent. The reduced solubility and increased chemical stability of L-cystine allow for its purification through crystallization. The resulting crystals are then dissolved in water and reduced back to L-cysteine using electrolysis [56].
More recently, Wacker AG (Munich, Germany) has developed a method for the direct purification of L-cysteine from a clarified fermentation broth [55]. This approach selectively isolates L-cysteine while eliminating its derivatives, emphasizing the importance of preventing the oxidation of L-cysteine to L-cystine. The oxidation of L-cysteine’s sulfhydryl (SH) groups by oxygen, the primary oxidizing agent present in fermentation broths, is particularly favored under alkaline conditions.
4. Conclusions
In general, the most widely researched microorganism for L-cysteine bioproduction is E. coli. Nonetheless, other production hosts like C. glutamicum and P. ananatis have been the focus of strain optimization, with promising improvements in L-cysteine production.
The combination of three metabolic engineering strategies has been successful, irrespective of the production host: The first one is enhancing the L-cysteine biosynthesis by overexpressing the genes encoding for feedback-insensitive L-serine O-acetyltransferase (SAT) and O-acetyl-L-serine sulfhydrylase. In E. coli, the feedback inhibition of SAT was bypassed through the introduction of an SAT from Arabidopsis thaliana. C. glutamicum showed a high L-cysteine production with the homologous overexpression of the cysE and serA genes.
The second metabolic engineering strategy is to reduce L-cysteine degradation by the microorganisms. Therefore, the L-cysteine desulfhydrase gene aecD was deleted in C. glutamicum, which increased the production of L-cysteine. In E. coli, the gene products of tnaA and metC were identified to catalyze the degradation of L-cysteine, and after the deletion of these genes, the L-cysteine production was improved.
The third metabolic engineering strategy is to enhance the export of L-cysteine and weaken the import. As L-cysteine is cytotoxic, it has to be transported out of the cell. In E. coli, two membrane proteins called YdeD and YfiK are responsible for the export of L-cysteine. But by overexpression of ydeD, OAS is simultaneously secreted, which causes the L-cysteine regulon activation to stop. In contrast, the gene product of yfiK enables L-cysteine export without depleting the intracellular OAS concentrations. In addition, the gene product of yeaN is a recently discovered L-cysteine uptake protein found in E. coli, which may be a promising target to avoid the reconsumption of L-cysteine by the cells.
Taking into account the process engineering aspects beyond metabolic engineering of the L-cysteine biosynthesis pathway enables economic L-cysteine production by fermentation. Emphasizing energy-efficient sulfur pathways, selecting economically viable carbon sources, designing scalable fed-batch processes with individual feedings of carbon and sulfur sources, and implementing efficient purification techniques are important for the production of L-cysteine on an industrial scale. The utilization of thiosulfate instead of sulfates requires fewer reduction equivalents, and thus, enhances L-cysteine production rates. Even though the highest L-cysteine concentrations are still obtained with glucose as a carbon source, microbial L-cysteine production with glycerol has been shown. Glycerol is abundant as a by-product of the biodiesel industry, and thus, its low cost might improve the industrial-scale L-cysteine production in the future.
References
- Paul, B.D.; Sbodio, J.I.; Snyder, S.H. Cysteine metabolism in neuronal redox homeostasis. Trends Pharmacol. Sci. 2018, 39, 513–524.
- Kredich, N.M. Regulation of l-Cysteine Biosynthesis in Salmonella typhimurium: I. Effects of growth on varying sulfur sources and O-acetyl-L-serine on gene expression. J. Biol. Chem. 1971, 246, 3474–3484.
- Sørensen, M.A.; Pedersen, S. Cysteine, even in low concentrations, induces transient amino acid starvation in Escherichia coli. J. Bacteriol. 1991, 173, 5244–5246.
- Takumi, K.; Nonaka, G. Bacterial Cysteine-Inducible Cysteine Resistance Systems. J. Bacteriol. 2016, 198, 1384–1392.
- Yin, J.; Ren, W.; Yang, G.; Duan, J.; Huang, X.; Fang, R.; Li, C.; Li, T.; Yin, Y.; Hou, Y.; et al. L-Cysteine metabolism and its nutritional implications. Mol. Nutr. Food Res. 2016, 60, 134–146.
- Takumi, K.; Ziyatdinov, M.K.; Samsonov, V.; Nonaka, G. Fermentative production of cysteine by Pantoea ananatis. Appl. Environ. Microbiol. 2017, 83, e02502-16.
- Wei, L.; Wang, H.; Xu, N.; Zhou, W.; Ju, J.; Liu, J.; Ma, Y. Metabolic engineering of Corynebacterium glutamicum for l-cysteine production. Appl. Microbiol. Biotechnol. 2019, 103, 1325–1338.
- Wendisch, V.F. Metabolic engineering advances and prospects for amino acid production. Metab. Eng. 2020, 58, 17–34.
- Clemente Plaza, N.; Reig García-Galbis, M.; Martínez-Espinosa, R.M. Effects of the Usage of l-Cysteine (l-Cys) on Human Health. Molecules 2018, 23, 3.
- Ismail, N.I.; Yumi Zuhanis Has-Yun, H.; Parveen, J. Production of Cysteine: Approaches, Challenges and Potential Solution. Int. J. Biotechnol. Wellness Ind. 2014, 3, 95–101.
- Bichon, E.; Sitthisack, P.; Sérée-Vattier, L.; Prévost, S.; Schiphorst, A.-M.; Pouponneau, K.; Monteau, F.; Le Bizec, B. Determination of l-cysteine origin on the basis of its δ15N values. Food Chem. 2018, 260, 283–288.
- Takagi, H.; Ohtsu, I. L-Cysteine Metabolism and Fermentation in Microorganisms. In Amino Acid Fermentation; Yokota, A., Ikeda, M., Eds.; Springer: Tokyo, Japan, 2017; pp. 129–151.
- Liu, H.; Fang, G.; Wu, H.; Li, Z.; Ye, Q. L-Cysteine Production in Escherichia coli Based on Rational Metabolic Engineering and Modular Strategy. Biotechnol. J. 2018, 13, 1700695.
- Sano, K.; Yokozeki, K.; Tamura, F.; Yasuda, N.; Noda, I.; Mitsugi, K. Microbial conversion of DL-2-amino-delta2-thiazoline-4-carboxylic acid to L-cysteine and L-cystine: Screening of microorganisms and identification of products. Appl. Environ. Microbiol. 1977, 34, 806–810.
- Sano, K.; Mitsugi, K. Enzymatic Production of l-Cysteine from dl-2-Amino-Δ2-thiazoline-4-carboxylic Acid by Pseudomonas thiazolinophilum: Optimal Conditions for the Enzyme Formation and Enzymatic Reaction. Agric. Biol. Chem. 1978, 42, 2315–2321.
- Tamura, Y.; Nishino, M.; Ohmachi, T.; Asada, Y. N-Carbamoyl-L-Cysteine as an Intermediate in the Bioconversion from D,L-2-Amino-Δ2-Thiazoline-4-Carboxylic Acid to L-Cysteine by Pseudomonas sp. ON-4a. Biosci. Biotechnol. Biochem. 1998, 62, 2226–2229.
- Shiba, T.; Takeda, K.; Yajima, M.; Tadano, M. Genes from Pseudomonas sp. Strain BS Involved in the Conversion of l-2-Amino-Δ2-Thiazolin-4-Carbonic Acid to l-Cysteine. Appl. Environ. Microbiol. 2002, 68, 2179–2187.
- Huai, L.; Chen, N.; Yang, W.; Bai, G. Metabolic control analysis of L-cysteine producing strain TS1138 of Pseudomonas sp. Biochemistry 2009, 74, 288–292.
- Wendisch, V.F. Microbial production of amino acids and derived chemicals: Synthetic biology approaches to strain development. Curr. Opin. Biotechnol. 2014, 30, 51–58.
- Pizer, L.I. The pathway and control of serine biosynthesis in Escherichia coli. J. Biol. Chem. 1963, 238, 3934–3944.
- Vermeij, P.; Kertesz, M.A. Pathways of assimilative sulfur metabolism in Pseudomonas putida. J. Bacteriol. 1999, 181, 5833–5837.
- Ono, B.-I.; Hazu, T.; Yoshida, S.; Kawato, T.; Shinoda, S.; Brzvwczy, J.; Paszewski, A. Cysteine biosynthesis in Saccharomyces cerevisiae: A new outlook on pathway and regulation. Yeast 1999, 15, 1365–1375.
- Sperandio, B.; Polard, P.; Ehrlich, D.S.; Renault, P.; Guédon, E. Sulfur amino acid metabolism and its control in Lactococcus lactis IL1403. J. Bacteriol. 2005, 187, 3762–3778.
- Byrne, C.R.; Monroe, R.S.; Ward, K.A.; Kredich, N.M. DNA sequences of the cysK regions of Salmonella typhimurium and Escherichia coli and linkage of the cysK regions to ptsH. J. Bacteriol. 1988, 170, 3150–3157.
- Phue, J.-N.; Shiloach, J. Transcription levels of key metabolic genes are the cause for different glucose utilization pathways in E. coli B (BL21) and E. coli K (JM109). J. Biotechnol. 2004, 109, 21–30.
- Tanaka, N.; Hatano, T.; Saito, S.; Wakabayashi, Y.; Abe, T.; Kawano, Y.; Ohtsu, I. Generation of hydrogen sulfide from sulfur assimilation in Escherichia coli. J. Gen. Appl. Microbiol. 2019, 65, 234–239.
- Liu, H.; Hou, Y.; Wang, Y.; Li, Z. Enhancement of Sulfur Conversion Rate in the Production of l-Cysteine by Engineered Escherichia coli. J. Agric. Food Chem. 2020, 68, 250–257.
- Liu, H.; Wang, Y.; Hou, Y.; Li, Z. Fitness of Chassis Cells and Metabolic Pathways for l -Cysteine Overproduction in Escherichia coli. J. Agric. Food Chem. 2020, 68, 14928–14937.
- Nakamori, S.; Kobayashi, S.I.; Kobayashi, C.; Takagi, H. Overproduction of L-cysteine and L-cystine by Escherichia coli strains with a genetically altered serine acetyltransferase. Appl. Environ. Microbiol. 1998, 64, 1607–1611.
- Takagi, H.; Kobayashi, C.; Kobayashi, S.; Nakamori, S. PCR random mutagenesis into Escherichia coli serine acetyltransferase: Isolation of the mutant enzymes that cause overproduction of L-cysteine and L-cystine due to the desensitization to feedback inhibition. FEBS Lett. 1999, 452, 323–327.
- Takagi, H.; Awano, N.; Kobayashi, S.; Noji, M.; Saito, K.; Nakamori, S. Overproduction of l-cysteine and l-cystine by expression of genes for feedback inhibition-insensitive serine acetyltransferase from Arabidopsis thaliana in Escherichia coli. FEMS Microbiol. Lett. 1999, 179, 453–459.
- Wirtz, M.; Hell, R. Production of cysteine for bacterial and plant biotechnology: Application of cysteine feedback-insensitive isoforms of serine acetyltransferase. Amino Acids 2003, 24, 195–203.
- Noji, M.; Inoue, K.; Kimura, N.; Gouda, A.; Saito, K. Isoform-dependent Differences in Feedback Regulation and Subcellular Localization of Serine Acetyltransferase Involved in Cysteine Biosynthesis from Arabidopsis thaliana. J. Biol. Chem. 1998, 273, 32739–32745.
- Hirasawa, T.; Shimizu, H. Glutamic Acid Fermentation: Discovery of Glutamic Acid-Producing Microorganisms, Analysis of the Production Mechanism, Metabolic Engineering, and Industrial Production Process. In Industrial Biotechnology; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; pp. 339–360.
- Kinoshita, S.; Udaka, S.; Shimono, M. Studies on the Amino Acid Fermentation. J. Gen. Appl. Microbiol. 1957, 3, 193–205.
- Joo, Y.-C.; Hyeon, J.E.; Han, S.O. Metabolic Design of Corynebacterium glutamicum for Production of l-Cysteine with Consideration of Sulfur-Supplemented Animal Feed. J. Agric. Food Chem. 2017, 65, 4698–4707.
- Mundhada, H.; Schneider, K.; Christensen, H.B.; Nielsen, A.T. Engineering of high yield production of L-serine in Escherichia coli. Biotechnol. Bioeng. 2016, 113, 807–816.
- Becker, J.; Wittmann, C. Bio-based production of chemicals, materials and fuels—Corynebacterium glutamicum as versatile cell factory. Curr. Opin. Biotechnol. 2012, 23, 631–640.
- Park, J.H.; Lee, S.Y. Towards systems metabolic engineering of microorganisms for amino acid production. Curr. Opin. Biotechnol. 2008, 19, 454–460.
- Wada, M.; Awano, N.; Haisa, K.; Takagi, H.; Nakamori, S. Purification, characterization and identification of cysteine desulfhydrase of Corynebacterium glutamicum, and its relationship to cysteine production. FEMS Microbiol. Lett. 2002, 217, 103–107.
- Rossol, I.; Pühler, A. The Corynebacterium glutamicum aecD gene encodes a C-S lyase with alpha, beta-elimination activity that degrades aminoethylcysteine. J. Bacteriol. 1992, 174, 2968–2977.
- Awano, N.; Wada, M.; Kohdoh, A.; Oikawa, T.; Takagi, H.; Nakamori, S. Effect of cysteine desulfhydrase gene disruption on l-cysteine overproduction in Escherichia coli. Appl. Microbiol. Biotechnol. 2003, 62, 239–243.
- Kawano, Y.; Ohtsu, I.; Takumi, K.; Tamakoshi, A.; Nonaka, G.; Funahashi, E.; Ihara, M.; Takagi, H. Enhancement of l-cysteine production by disruption of yciW in Escherichia coli. J. Biosci. Bioeng. 2015, 119, 176–179.
- Tchong, S.-I.; Xu, H.; White, R.H. l-Cysteine Desulfidase: An Enzyme Isolated from Methanocaldococcus jannaschii That Catalyzes the Breakdown of l-Cysteine into Pyruvate, Ammonia, and Sulfide. Biochemistry 2005, 44, 1659–1670.
- Daßler, T.; Maier, T.; Winterhalter, C.; Böck, A. Identification of a major facilitator protein from Escherichia coli involved in efflux of metabolites of the cysteine pathway. Mol. Microbiol. 2000, 36, 1101–1112.
- Pittman, M.S.; Corker, H.; Wu, G.; Binet, M.B.; Moir, A.J.G.; Poole, R.K. Cysteine Is Exported from the Escherichia coli Cytoplasm by CydDC, an ATP-binding Cassette-type Transporter Required for Cytochrome Assembly *. J. Biol. Chem. 2002, 277, 49841–49849.
- Franke, I.; Resch, A.; Dassler, T.; Maier, T.; Böck, A. YfiK from Escherichia coli promotes export of O-acetylserine and cysteine. J. Bacteriol. 2003, 185, 1161–1166.
- Caballero Cerbon, D.A.; Widmann, J.; Weuster-Botz, D. Metabolic control analysis enabled the improvement of the L-cysteine production process with Escherichia coli. Appl. Microbiol. Biotechnol. 2024, 108, 1–13.
- Nakatani, T.; Ohtsu, I.; Nonaka, G.; Wiriyathanawudhiwong, N.; Morigasaki, S.; Takagi, H. Enhancement of thioredoxin/glutaredoxin-mediated L-cysteine synthesis from S-sulfocysteine increases L-cysteine production in Escherichia coli. Microb. Cell Factories 2012, 11, 62.
- Kawano, Y.; Onishi, F.; Shiroyama, M.; Miura, M.; Tanaka, N.; Oshiro, S.; Nonaka, G.; Nakanishi, T.; Ohtsu, I. Improved fermentative l-cysteine overproduction by enhancing a newly identified thiosulfate assimilation pathway in Escherichia coli. Appl. Microbiol. Biotechnol. 2017, 101, 6879–6889.
- Kredich, N.M.; Tomkins, G.M. The enzymic synthesis of L-cysteine in Escherichia coli and Salmonella typhimurium. J. Biol. Chem. 1966, 241, 4955–4965.
- Kawano, Y.; Suzuki, K.; Ohtsu, I. Current understanding of sulfur assimilation metabolism to biosynthesize l-cysteine and recent progress of its fermentative overproduction in microorganisms. Appl. Microbiol. Biotechnol. 2018, 102, 19.
- Zhu, Z.; Zhang, J.; Ji, X.; Fang, Z.; Wu, Z.; Chen, J.; Du, G. Evolutionary engineering of industrial microorganisms-strategies and applications. Appl. Microbiol. Biotechnol. 2018, 102, 4615–4627.
- Zhang, X.; Zhang, D.; Zhu, J.; Liu, W.; Xu, G.; Zhang, X.; Shi, J.; Xu, Z. High-yield production of l-serine from glycerol by engineered Escherichia coli. J. Ind. Microbiol. Biotechnol. 2019, 46, 221–230.
- Boehm, A. Process for Purifying L-Cysteine. U.S. Patent US8088949B2, 3 January 2012. Available online: https://patents.google.com/patent/US8088949B2/en (accessed on 22 September 2023).
- Doering, W. Verfahren zur Isolierung von Cystin. German Patent DE10040176A1, 8 September 2002. Available online: https://patents.google.com/patent/DE10040176A1/en/en17 (accessed on 13 September 2023).
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
630
Revisions:
2 times
(View History)
Update Date:
23 Jan 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





