Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Nafees Khan | -- | 4524 | 2024-01-18 16:01:47 | | | |
| 2 | Lindsay Dong | Meta information modification | 4524 | 2024-01-19 04:21:03 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Khan, S.; Alvi, A.F.; Khan, N.A. Role of Ethylene in Plant Developmental Processes. Encyclopedia. Available online: https://encyclopedia.pub/entry/54059 (accessed on 08 February 2026).
Khan S, Alvi AF, Khan NA. Role of Ethylene in Plant Developmental Processes. Encyclopedia. Available at: https://encyclopedia.pub/entry/54059. Accessed February 08, 2026.
Khan, Sheen, Ameena Fatima Alvi, Nafees A. Khan. "Role of Ethylene in Plant Developmental Processes" Encyclopedia, https://encyclopedia.pub/entry/54059 (accessed February 08, 2026).
Khan, S., Alvi, A.F., & Khan, N.A. (2024, January 18). Role of Ethylene in Plant Developmental Processes. In Encyclopedia. https://encyclopedia.pub/entry/54059
Khan, Sheen, et al. "Role of Ethylene in Plant Developmental Processes." Encyclopedia. Web. 18 January, 2024.
Copy Citation
Ethylene, a gaseous phytohormone, is emerging as a central player in the intricate web of plant developmental processes from germination to senescence under optimal and stressed conditions. The presence of ethylene has been noted in different plant parts, including the stems, leaves, flowers, roots, seeds, and fruits.
ethylene
plant
cell division and elongation
flower and fruit development
chloroplast development
photosynthesis
senescence and abscission
1. Introduction
Plant development is influenced by several intrinsic and extrinsic factors that coordinate to regulate all processes in the life cycles of plants. Plant growth is one carefully regulated process that occurs throughout the vegetative phase. Plant growth regulators act as regulators of plant developmental processes and are crucial for the resilience of responses under stress. Ethylene (ET) is a versatile hormone involved in development, metabolism, and stress responses in plants. Researchers have shown manifold roles of this hormone in the development of plants, as a signaling agent, in leaf development, in senescence, in fruit ripening, in the promotion of germination, etc. ET is the second most fundamental unsaturated hydrocarbon. It exhibits peculiar dose-dependent actions under ideal and external disruptions for all its suppression or promotion responses like germination, ripening, growth, and senescence. For instance, a low concentration of ET facilitates the activation of defense signaling in plants; its high concentration seems to inhibit development in Triticum aestivum and Cucumis sativus [1][2][3]. Plants growing in compacted soil experience excessive accumulation and concentration of ET in root tissues, resulting in root growth inhibition [4]. Increasing evidence confirms that ET is crucial in plant architecture. In Zea mays, ACS7 (1-aminocyclopropane-1-carboxylic acid synthases 7) mutation brought about a dwarf phenotype with a larger leaf angle on Sdw3 (semi-dwarf3) due to increased ET synthesis [5].
ET plays a central role in the growth and development process, starting from seed germination to senescence. However, complex crosstalk between phytohormones like abscisic acid (ABA), gibberellin (GA), cytokinin, and auxin regulates the responses sometimes antagonistically or synergistically [6][7]. ET biosynthesis or signaling mutant showed differential responses to hormonal sensitivity and plant developmental processes. For example, ethylene overproduction 3 (eto3) and constitutive triple response 1 (ctr1) mutants correspond to ABA insensitivity, whereas ethylene receptor 1 (etr1), ethylene-insensitive 2 (ein2), and ethylene-insensitive 6 (ein6) show enhancement of ABA sensitivity [8]. The faulty hook formation in ET-insensitive mutant ein2 signifies the prominence of ET-mediated auxin biosynthesis for hook development and regulation [9]. Moreover, EIN3 synchronizes the expression of chlorophyll biosynthesis genes PORA/B (PROTOCHLOROPHYLLIDE OXIDOREDUCTASE A/B) [10]: the pigment-binding proteins LHC which are essential for photosynthesis initiation. EIN3/EIL1 chiefly regulates ethylene signaling responses, thereby tuning the range of transcriptional regulation depending on spatiotemporal and environmental conditions [11]. Exogenous application of ET has been found to increase the number of lateral roots, root fresh weight, and mineral content in the root and shoot system, along with the upregulation of auxin biosynthesis and transportation genes [12]. In addition, ET has a significant effect on flower development and sex differentiation. ACS gene family has been widely studied and characterized for its potential function in sex determination [13] Fruit ripening is another ET-dependent process associated with multiple biological events like respiration, pigment accumulation, ET production, change in texture, and overall building up of fruit quality traits. ET activity participates in gene expression responsible for the aforementioned changes during ripening [14].
2. Ethylene Biosynthesis and Signaling
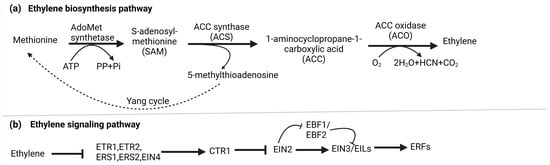
Ethylene biosynthesis involves a two-step enzymatic pathway starting with methionine, which is converted into S-adenosyl-L-methionine (SAM or S-AdoMet) by S-AdoMet synthetase. This transformation utilizes one ATP molecule. Subsequently, S-AdoMet is enzymatically converted into 1-aminocyclopropane-1-carboxylic acid (ACC) and 5′-methylthioadenosine (MTA) by ACC synthase (ACS) [15]. Lastly, ET is generated through the oxidation of ACC, facilitated by ACC oxidase (ACO). In parallel, the Yang cycle operates to convert MTA, a byproduct of the second stage, back into methionine, ensuring an optimal methionine pool [16]. Under normal basal ET production levels, ACS is believed to control the rate-limiting step in biosynthesis [15]. Nevertheless, specific circumstances can lead to ACO becoming the limiting factor [17]. Figure 1a illustrates the biosynthesis of ET.

Figure 1. (a) Biosynthesis of ethylene: In higher plants, ethylene is produced through the Methionine/Yang cycle. This involves converting methionine into 1-aminocyclopropane-1-carboxylic acid (ACC) using ACC synthase (ACS), and then ACC is further converted into ethylene by ACC oxidase (ACO). The Yang cycle recycles 5-methylthioadenosine to regenerate methionine, which is crucial for this process. (b) Signaling pathway of ethylene: Ethylene receptors (ETR1, ERS1, EIN4, ETR2, and ERS2) on the endoplasmic reticulum (ER) activate CTR1 kinase, which phosphorylates and degrades EIN2s C-terminal part, preventing its nucleus entry. Simultaneously, EBF1/2 transcripts produce F-box proteins that target EIN3/EIL transcription factors for degradation, blocking ethylene response. Ethylene binding deactivates CTR1, leading to EIN2 dephosphorylation. A protease cleaves EIN2s C-terminal part (EIN2-CEND) in the nucleus and cytoplasm. EIN2-CEND then enters the nucleus and binds to EIN3/EIL transcription factors, initiating gene expression for ethylene response.
The canonical ethylene signaling pathway begins when ethylene binds to ER-localized receptor proteins, such as ETHYLENE RESPONSE 1 (ETR1), ETHYLENE RESPONSE 2 (ETR2), ETHYLENE-INSENSITIVE 4 (EIN4), ETHYLENE RESPONSE SENSOR 1 (ERS1), and ERS2 in Arabidopsis, organized into two subfamilies defined by their ET binding and histidine kinase domains. Without ET, these receptors activate CONSTITUTIVE TRIPLE RESPONSE (CTR1), a Ser-Thr protein kinase on the ER membrane, which phosphorylates ETHYLENE-INSENSITIVE 2 (EIN2) C-terminal end (EIN2-CEND). F-box proteins ETP1 and ETP2 target phosphorylated EIN2 for degradation by the 26 S proteasome, preventing its nuclear translocation [18].
3. Snapshot of Regulation of Ethylene Biosynthesis
Since ACS and ACO are the exclusive enzymes involved in ET biosynthesis, most regulation concerning overall ET production revolves around modulating these pivotal enzymes’ transcription, translation, and protein stability. However, these enzymes are also affected by phytohormones and other stimuli like light and stress. In the early stages of research on the transcriptional regulation of ET biosynthesis, scientists uncovered the presence of a multigene family known as ACS genes, which exhibited distinct expression patterns in plants. Notably, four specific ACS genes in S. lycopersicum were pivotal in orchestrating the shift from autoinhibitory to autocatalytic ET production during fruit ripening [19]. Leading the charge as the initial regulator of ACS expression was the MADS-box transcription factor SlRIN, which directly bolstered the expression of selected S. lycopersicum ACS genes [20]. Over the years, numerous other transcription factors have emerged as key players, promoters, and inhibitors of ACS gene expression, influencing diverse growth processes. However, there is also supporting information suggesting that ET itself can directly influence ACS transcription. For instance, the ET response factor SlERF2/TERF2 of S. lycopersicum interacts with the promoter of NtACS3, thereby stimulating its expression [21]. The regulation of ET synthesis is further complicated by its interaction with light, phytohormones, and various biotic or abiotic stresses. The impact of light on ACS gene expression varies, contingent on developmental stages and light conditions. Notably, different ACS isozymes seem to have specific functions, with ACS5, 6, 8, and 9 primarily influencing ET production in dark-grown seedlings, while ACS2 and ACS4 assume control over ET production in well-lit conditions [22]. Mutations in the phytochrome genes PHYA and PHYB have been found to affect ET biosynthesis, with phyA mutants displaying a more pronounced increase in ET production.
Much like their ACS counterparts, ACO genes are subject to transcriptional regulation. Notably, the S. lycopersicum HD-ZIP transcription factor SlHB-1 and the ripening regulator RIN directly stimulate ACO gene expression. Across various species, different classes of transcription factors have also been shown to regulate ACO. Interestingly, ET can directly control ACO expression through intricate feedback mechanisms mediated by ERF (ethylene response factor) proteins [23]. In S. lycopersicum, ACO1 experiences upregulation in response to white light pulses. This phenomenon propels ET production and serves as a reporting mechanism for ET responses, thus establishing a positive feedback loop. Additionally, ACO transcript levels surge after ACC treatment of light-grown seedlings. During phases of ripening or stress, when ACS activity is maximum, ACO activity becomes rate-limiting, prompting an upswing in ACO expression. This orchestrated feed-forward mechanism is pivotal in eliminating excess ACC when ACO activity imposes restrictions on ET production [22].
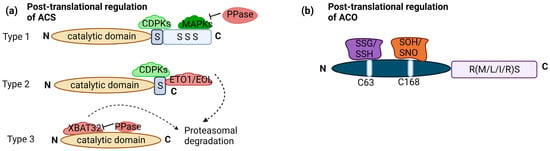
Transcriptional regulation of ET biosynthesis is complemented by posttranslational modifications of ACS proteins. ACS proteins share a conserved N terminus and catalytic core, there’s notable variability in the C terminus among different ACS isoforms. This variability leads to the classification of ACS proteins into three major groups in Arabidopsis: type 1 (ACS1, 2 and 6), type 2 (ACS4, 5, 8, 9, and 11), and type 3 (ACS7). Type 1 has a phosphorylation site for mitogen-activated protein kinases (MAPKs) and calcium-dependent protein kinases (CDPKs). Type 2 has a site for CDPKs and ETO1 (Ethylene Overproducer1)/ETO1-LIKE1 (EOL), an E3 ligase leading to degradation. Type 3 has no target site. Phosphorylation stabilizes type 1 ACS, promoting ET production. Conversely, dephosphorylation can lead to ACS degradation, although this effect depends on the type of ACS [23]. Moreover, phosphorylation can destabilize type 2 ACS, as exemplified by the phosphorylation of ACS5 (type 2) by Casein Kinase 1.8 (CK1.8), resulting in E3 ubiquitin ligases mediated degradation [24]. Light-triggered posttranslational regulation of type-2 ACSs, especially ACS5, regulating hypocotyl elongation during the dark-to-light shift [25]. PIF3 plays a key role, with ET stabilizing PIF3 in light. This light-stimulated stabilization of ACS enzymes increases ET production, potentially contributing to PIF3 stability and the subsequent promotion of ET-driven hypocotyl elongation under light exposure [26]. PP2A, a regulatory component, plays a role in posttranslational ACS stability regulation. PP2A-mediated dephosphorylation negatively affects ACS6 protein stability in the dark [27]. Posttranslational regulation of ACS also involves ubiquitination, with type 3 ACS7 being targeted for degradation through ubiquitination mediated by XBAT32 (XB3 orthologue 2 in Arabidopsis (Figure 2a). Protein Phosphatase 2C family members (PP2C’s) also play a role in stabilizing ACS7 [23].

Figure 2. Posttranslational regulation of ACS and ACO: (a) ACS enzymes can be categorized into three groups based on their phosphorylation sites: type 1 with both CDPK (calcium-dependent protein kinase) and MAPK (Mitogen-activated protein kinase) sites, type 2 with only CDPK sites, and type 3 with no C-terminal regulatory sites. Type 1 ACS is positively regulated by MAPKs and CDPKs, significantly boosting its activity and ethylene production. Phosphatases (PPase) are involved in dephosphorylation, negatively impacting protein stability. Type 2 ACSs are regulated by the ETO1-containing E3 ligase, recognizing the TOE (Target of ETO1) domain in their C-termini. CDPK phosphorylation likely plays a role in regulating this ubiquitination process and, thus, the stability of Type 2 ACS protein. XBAT32 directly binds to type 3 ACS, leading to protein degradation, while PPase plays a role in stabilizing. (b) ACO enzymes fall into three related groups based on amino acids in the RXS motif: type 1 with RMS intermediate residue, type 2 with R-L/I-S intermediate residue, and type 3 with RRS intermediate residue. Type 1 ACOs can undergo modifications at C63, including S-glutathionylation (SSG) and S-sulfhydration (SSH), as well as at C168, including S-sulfenylation (SOH) and S-nitrosylation (SNO). No such modifications have been observed in type 2 and type 3 ACOs. The figure is based on and modified from [23]. Dotted arrow—multistep pathway; solid arrow—promotion; flat-head arrow—inhibition.
While much is known about the posttranslational regulation of ACS, research on the posttranslational regulation of ACO has been comparatively limited. It is worth mentioning that the three categories of ACO can be distinguished based on the specific intermediate amino acid present within the conserved RXS motif. Type 1 ACOs contain an RMS intermediate residue, type 2 ACOs possess an R-L/I-S intermediate residue, and type 3 ACOs feature an RRS intermediate residue. ACO proteins undergo redox-specific posttranslational modifications. These modifications involve cysteine residues and include S-glutathionylation (SSG), S-sulfhydration (SSH) at cysteine (C63), S-sulfenylation (SOH), and S-nitrosylation (SNO) at C168. These modifications are recorded only in type 1 ACO (Figure 2b).
4. Involvement of Ethylene in Plant Developmental Processes
4.1. Cell Division and Elongation
The role of ET in cell division is complex. Ethylene affects cell division depending on the specific tissue type and the internal and external signals at play. In certain situations, ET acts as a stimulator of cell division. For example, during the development of the apical hook and during the early stages of apical hook development, ethylene appears to play a synergistic role with auxins in stimulating cell division within the subepidermal layers. This collaborative effect is crucial for the bending of the apical hook.
However, the role of ET in cell division is not uniform across all plant tissues. Ethylene’s effects on cell division in the root system are somewhat contradictory. Studies have shown that ET does not significantly alter the expression pattern of CYCLIN-DEPENDENT PROTEIN KINASE B1;1 (CYCB1;1), pointing out that it does not directly affect mitotic activity in the root. However, it is important to note that ET modulates cell division within the quiescent center, developing additional columella layers in the root cap [28][29].
The impact of ET on leaf cell division is context-dependent. Under environmental stress, particularly when plants experience less than 10 h of osmotic stress, ET mediates transient and reversible cell cycle cessation. This effect is thought to involve the phosphorylation-mediated inactivation of CYCLIN-DEPENDENT KINASE A (CDKA), possibly via the MPK3/6 pathway. Notably, this cell cycle arrest operates independently of the EIN3 transcriptional control [30]. Cell cycle inhibition by ET in leaves is multifaceted; ET accumulation activates BOLITA, an ERF, activating type II TCP (TEOSINTE BRANCHED 1/CYCLOIDEA/PCF) genes [31]. These TCP proteins then bind to the promoter of RBR1 (RETINOBLASTOMA RELATED 1), phosphorylating E2Fa and repressing E2F target gene transcription, thereby impeding progression into the S-phase and cell division [32].
Cellular growth relies on key processes like the rearrangement of the cytoskeleton, the modification of the cell wall facilitated by cell-wall-remodeling enzymes, and water uptake via aquaporins. Cell elongation involves rearranging cortical microtubules (CMTs) perpendicular to the growth axis [33]. Ethylene rapidly alters CMT orientation in Arabidopsis roots and hypocotyls, inhibiting cell elongation and promoting radial swelling within 10 min [33].
4.2. Leaf Growth and Flower Development
Ethylene overproduction in Arabidopsis resulted in dwarfed plants with reduced growth [34][35]. Accordingly, when positive regulators of the ET signaling pathway in plants were mutated, these plants exhibited rosette leaves compared to control plants. Mutation of the endoplasmic reticulum (ER)-anchored protein EIN2, for instance, has been linked to increased growth [36]. On the contrary, mutations in components that inhibited ET signaling, like the receptors ETR1 and ERS1, resulted in stunted growth.
The regulatory mechanism of sex determination in plants represents a model experiment system in the case of unisexual plants [37]. Considerable focus has been directed towards identifying genes responsible for regulating the development of male and female flowers. Although various plant hormones were investigated for their impact on the proportion of unisexual flowers, ET emerged as a prominent regulator of unisexual flower development [38]. The primary genes determining flower sex type encode crucial enzymes engaged in ethylene production, with Wiskott–Aldrich syndrome protein-interacting protein (WIP1), which encodes a C2H2 zinc finger transcription factor of the WIP family. The M (Monoecious) gene encodes ACS2, expressed in the carpel region of the female flower; its inactivation (m) leads to the development of a bisexual flower [39]. ACO2 has also been demonstrated to impact the formation of unisexual flowers by collaborating with ACS11 to promote the selective production of ET in the floral region; dysfunctional ACO2 results in the absence of female flowers [40]
4.3. Root Hair Development
Root hairs represent an extensive network of epidermal cells within the root system, playing a vital role in nutrient uptake, anchoring the plant in the soil, and facilitating interactions with the environment in stationary plants. The plant hormone ET not only fosters the growth of these root hairs but also acts as a mediator for various signals that trigger the development of hair cells [41]. Ethylene’s role in Arabidopsis root hair formation is elucidated through ET biosynthesis mutants. Root hairs of eto1, eto2, and eto4 mutants develop longer hairs than wild-type ones on trichoblasts cells. Interestingly, the eto3 mutant stands out as it produces higher levels of ET than other eto alleles. Remarkably, it also triggers the development of hairs on atrichoblasts, which are typically hairless cells [42]. Mutations in genes involved in ET signaling confirm ET necessity for root hair elongation; for example, etr1 and ein2 receptor mutations yield shorter root hairs (50–70% of wild type), while ctr1 loss-of-function mutations produce longer root hairs. This underscores ET’s crucial role in controlling root hair length [42].
4.4. Fruit Ripening
Ethylene plays a crucial role in the regulation of fruit ripening by orchestrating the expression of genes involved in various biological processes. These processes encompass pigment accumulation, respiration, ET production, texture change, and the overall enhancement of fruit quality traits [14]. Excess ET production is chiefly associated with transcriptional upregulation of ET biosynthesis genes (ACS and ACO). Until now, 14 ACS genes have been identified in the S. lycopersicum, among these, ACS2 and ACS4 showed important fruit-ripening functions [43]. Through genome-wide identification, six ACO genes have been identified in S. lycopersicum. Interestingly, during the pre-ripening stages, the expression levels of all ACO genes remain undetectable. However, ACO1 and ACO2 demonstrate high expression levels during the ripening process, while ACO4 exhibits a gradual and slight increase in expression. Upon ripening, the undetectable transcript level of ACO3, ACO5, and ACO6 suggests the least role in climacteric ET production [14]. During the climacteric stage, there is a well-known transition from system 1 to system 2, accompanied by a remarkable upregulation of ACS2, ACS4, ACO1, and ACO4. This upregulation leads to positive feedback regulation. In S. lycopersicum, the expressions of ACS1A, ACS3, ACS6, ACO1, and ACO4 are associated with system 1 of ethylene production, while the expressions of ACS2, ACS4, ACO1, and ACO4 are associated with system 2 [19][44].
Unlike climacteric fruits such as Solanum lycopersicum, non-climacteric fruits, such as Fragaria x ananassa (strawberry) do not rely on ET for the initiation and maintenance of the ripening process [45]. However, the measurement of ACC revealed that it is present in large quantities as strawberries progress towards the stages of fruit ripening. ACC accumulation in strawberry directly indicates higher expression of ACS genes. According to the expression analysis, the genes FaACS1 and FaACS26 exhibited ripening-specific expression in the receptacle tissues of strawberries. On the other hand, the genes FaACS17, FaACS21, FaACS19, and FaACS23 were considered to be achene-specific ripening-induced genes.
4.5. Chloroplast Development
When seeds start to sprout in conditions lacking light, they activate a specific developmental program called skotomorphogenesis. In the case of plants like Arabidopsis, which undergoes hypogeal germination (germination below the soil surface), an enhancement in the length of the hypocotyl (the embryonic stem) occurs due to cell elongation. This elongation helps push the cotyledons against the mechanical pressure of the soil, allowing them to reach the light source. In the course of the process, proplastids, the precursors to all plastids, undergo both proliferation and differentiation. This transformation leads them to become etioplasts within the cotyledon cells, thus preparing plants for photosynthetic apparatus during the dark-to-light transition [46]. Etioplasts have a semi-crystalline membrane cluster called the prolamellar body (PLB). This structure contains essential components for photosynthesis, including prothylakoid membranes, protochlorophyllide, and protochlorophyllide oxidoreductase, a light-dependent enzyme that transforms protochlorophyllide into chlorophyllide [46].
Ethylene’s regulatory functions hinge on two transcription factors, EIN3 and EIL1. During seedling emergence, the photomorphogenic regulator CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) senses light fluctuations, while ET processes mechanical stress cues, collaboratively influencing EIN3 protein levels. In the nucleus transcription of PhANGs (PHOTOSYNTHESIS-ASSOCIATED NUCLEAR GENE) is suppressed by PIFs in coordination with other protein EIN3. COP1, an E3 ligase, targets EBF1/2 for degradation, stabilizes PIF/EIN3, and degrades photomorphogenesis stimulating transcription factors like HY5 (ELONGATED HYPOCOTYL5) [47][48][49]. The perception of light by photoreceptors initiates chloroplast biogenesis and the shift toward photoautotrophic growth. Light causes a transformation in the structure of phytochrome (phy), converting it from an inactive state to an active nuclear form. This active form then initiates the degradation of the PIFs (PHYTOCHROME INTERACTING FACTORS) [49].
4.6. Photosynthesis
4.6.1. Stomatal Regulation
Stomata play a pivotal role in managing the exchange of gases between the leaf’s interior and the atmosphere by regulating turgor pressure in guard cells. These dynamic stomatal adjustments facilitate the precise assimilation of CO2 during photosynthesis, all while mitigating water loss through transpiration. ET’s regulation of stomatal responses is characterized by its tendency to manifest dual and sometimes conflicting roles [50]. Ethylene induces stomatal closure in wild Arabidopsis by triggering H2O2 synthesis in guard cells, this process relies on NADPH oxidase AtrbohF. ETR1 mediates ethylene and H2O2 signaling in guard cells via EIN2 and ARR2-dependent pathway(s), identifying AtrbohF as a key mediator of stomatal responses to ET [51]. Ethylene-insensitive mutants (etr1-1) of Arabidopsis showed a smaller number of stomata and reduced stomatal conductance indicating ET influences the stomatal development process and negatively affects stomatal conductance [52][53]. Also, another study showed that the application of exogenous ET enhances stomatal conductance, photosynthesis, and growth in Brassica juncea plants under optimal and deficient nitrogen fertilization [54].
4.6.2. Chlorophyll Content
According to Ceusters and Van de Poel [55], the impact of ET on photosynthesis is contingent upon the age of the leaves. ET directly regulates the photosynthetic process in younger, non-senescing leaves, while in mature leaves, its influence is more indirect, primarily driving leaf senescence. Studies on ethylene-insensitive mutants of Arabidopsis (etr1-1) and Nicotiana showed a decline in the overall photosynthetic capacity of young non-senescing leaves of the plant due to a reduction in the expression of crucial photosynthesis-associated genes like CAB (CHLOROPHYLL A/B-BINDING PROTEIN) and the small subunit of Rubisco [56][57][58]. Grbic and Bleecker [58] found that, in etr1-1 mutants, there was a delay in the initiation of leaf senescence, which correlated with a postponement in the activation of SAGs (SENESCENCE-ASSOCIATED GENES). Furthermore, they observed elevated expression levels in genes associated with photosynthesis. When non-senescing leaves were treated with external ET, it reduced the expression of CAB and a subsequent decrease in chlorophyll content. These findings suggest that ET negatively regulates photosynthesis in non-senescing leaves, implying that a basal level of ET production and perception is necessary for normal photosynthetic function.
4.6.3. Light Reaction
The process of photosynthesis transforms light energy into chemical energy. For this, absorption of light energy by PSII and PSI is essential to facilitate electron transport and reduction in CO2 in the chloroplast. But sometimes, excessive absorption can lead to photochemical damage due to excessive ROS generation. Plants employ protective mechanisms like non-photochemical quenching (NPQ) to prevent over-reduction in photosystems. Chen and Gallie [59] demonstrated that ET controls energy-dependent non-photochemical quenching in Arabidopsis by inhibiting the xanthophyll cycle. In the above study, Arabidopsis eto1-1 mutants (ethylene overproducing) exhibited reduced capacity to convert violaxanthin to zeaxanthin due to impaired violaxanthin de-epoxidase activity. This leads to elevated reactive oxygen species production and increased photosensitivity in response to high light in these plants. Analyzing the intricacies of chlorophyll fluorescence through pulse-amplitude modulation fluorimetry, Kim et al. [60] have shed light on the impact of ET signaling mutations on photosystem II (PSII) activity in Arabidopsis. Specifically, their study uncovers that ET-insensitive mutants (etr1-1) exhibit diminished PSII activity in comparison to their wild-type counterparts. Notably, etr1-1 mutant lines, which are often used for ET-related investigations, can carry a consequential secondary mutation in ACCUMULATION AND REPLICATION3 (a second mutation in etr1-1 mutant of Arabidopsis responsible for producing premature stop codon in ARC3), prompting the need for supplementary corroborative lines of evidence, particularly in photosynthesis research. Ethylene signaling mutants derived from the arc3 secondary mutation (etr1-1sg) also demonstrate reduced maximum quantum efficiency, prolonged chlorophyll fluorescence lifetime of PSII, and decreased quantum yield of PSII.
4.6.4. Dark Reaction
The available literature show that ET also controls the dark reaction of photosynthesis. Tholen et al. [56] demonstrated that as vegetative Nicotiana plants were cultivated under varying atmospheric CO2 concentrations, an inverse relationship was observed between glucose concentration within leaves and the expression of the Rubisco gene. This repression of gene expression was distinctly amplified by heightened glucose levels in plants insensitive to ethylene. The insensitivity to ET led to equivalent nitrogen allocations in light harvesting while experiencing diminished levels in electron transport and Rubisco. This, in turn, resulted in a noticeably diminished photosynthetic capacity in ethylene-insensitive transgenic Nicotiana plants compared to the wild type. These findings imply that the lack of ET perception enhanced the plants’ vulnerability to glucose, potentially due to escalated ABA concentrations. Ultimately, this increased susceptibility to endogenous glucose detrimentally affected Rubisco content and these plants’ carboxylation process and overall photosynthetic capacity. A similar decrease in the photosynthesis of Arabidopsis etr1 mutant was observed due to a decline in the content of Rubisco protein and expression level. Another study observed that overexpression of CitERF13 in tobacco leaves decreases the maximum rate of Rubisco carboxylase activity [61].
4.7. Senescence
Leaf senescence is a highly programmed, regulated, and degenerative process. It is characterized by chlorophyll breakdown and degradation of macromolecules [62]. Studies reported increased ET production in senescent leaves with higher transcription rates of ET biosynthesis genes ACS and ACO [63]. On the other hand, the Octuple mutant of ACS genes showed a delayed senescence response [64].
A study revealed that EIN2 plays a role in regulating leaf senescence partially via microRNA164 (miR164) and ORESARA1 (ORE1, also named ANAC092 or NAC2) [65]. A recent study showed that EIN3 and ORE1 can directly regulate the expression of CHLOROPHYLL CATABOLIC GENES (CCGs), NONYELLOWING1 (NYE1, also known as STAY-GREEN1, SGR1), NONYELLOW COLORING1 (NYC1), and PHEOPHORBIDE A OXYGENASE (PAO), thereby initiating chlorophyll breakdown during leaf senescence [66]. An ETHYLENE-INSENSITIVE3-LIKE1 (EIL1) gene, GhLYI (LINT YIELD INCREASING), encodes a truncated protein that regulates senescence by directly targeting SENESCENCE-ASSOCIATED GENE 20 (SAG20) in Gossypium hirsutum [67].
4.8. Abscission
The shedding of plant organs such as seedpods, leaves, floral organs, and fruits by detaching them at the abscission zone is called abscission [68]. Developmental and environmental changes can trigger abscission in plants and are mainly subjected to the crosstalk between two plant hormones, ET and auxin [69]. Ethylene regulates the gene expression pattern of enzymes involved in cell separation, like cellulases and pectinases [70]. Before abscission occurs, auxin is transported to the abscission zone to inhibit ET sensitivity in the cells. When abscission occurs, the organs undergoing abscission release ET, which is then followed by the detachment of leaves [71]. Ethylene present in the abscission zone initiates a signal transduction pathway that activates transcription factors and genes responsive to ET [72] which in turn, regulate abscission. In addition, the induction of ethylene production by methyl jasmonate was identified as the cause of fruit abscission in ‘Hamlin’ and ‘Valencia’ orange varieties [73]. For uniform ripening, external ET application promotes abscission in fruit crops. On the other hand, 2-aminoethoxyvinyl glycine (AVG), ET biosynthesis inhibitors are used to reduce abscission before harvest. Vesicle transport pathways genes in ethylene-induced AZ-C (calyx abscission zone) cells and adjacent FR (fruit rind) cells are responsible for citrus fruit abscission [74]. Hence, it appears that coordination between hydrolytic enzymes and ET production leads to plant organ abscission. The key ET biosynthetic enzymes ACS and ACO are found to be highly expressed during organ abscission, which facilitates ET production followed by activation of genes encoding cell-wall-remodeling enzymes [75][76]. Pollination upregulated the SlACS2 gene in S. lycopersicum [77]. In Petunia, pollination leads to a 20-fold increase in ET production autocatalytically from the stigmas contributing to wilting and eventually abscission [78].
5. Conclusions
In summary, ET is a remarkable and versatile plant hormone, wielding significant influence across a spectrum of plant developmental and physiological processes. It plays a pivotal role in a plant’s life cycle, including cell division, elongation, leaf growth, senescence, abscission, flower and fruit development, chloroplast maturation, and photosynthesis regulation. As our comprehension of ET involvement in plant development deepens, the potential for leveraging its properties to enhance crop performance and stress resilience becomes increasingly evident.
References
- Chang, C.; Wang, B.; Shi, L.; Li, Y.; Duo, L.; Zhang, W. Alleviation of salinity stress-induced inhibition of seed germination in cucumber (Cucumis sativus L.) by ethylene and glutamate. J. Plant Physiol. 2010, 167, 1152–1156.
- Khan, M.I.R.; Iqbal, N.; Masood, A.; Per, T.S.; Khan, N.A. Salicylic acid alleviates adverse effects of heat stress on photosynthesis through changes in proline production and ethylene formation. Plant Signal. Behav. 2013, 8, e26374.
- Riyazuddin, R.; Verma, R.; Singh, K.; Nisha, N.; Keisham, M.; Bhati, K.K.; Kim, S.T.; Gupta, R. Ethylene: A master regulator of salinity stress tolerance in plants. Biomolecules 2020, 10, 959.
- Pandey, B.K.; Huang, G.; Bhosale, R.; Hartman, S.; Sturrock, C.J.; Jose, L.; Martin, O.C.; Karady, M.; Voesenek, L.A.C.J.; Ljung, K.; et al. Plant roots sense soil compaction through restricted ethylene diffusion. Science 2021, 371, 276–280.
- Li, H.; Wang, L.; Liu, M.; Dong, Z.; Li, Q.; Fei, S.; Xiang, H.; Liu, B.; Jin, W. Maize plant architecture is regulated by the ethylene biosynthetic gene ZmACS7. Plant Physiol. 2020, 183, 1184–1199.
- Ahammed, G.J.; Gantait, S.; Mitra, M.; Yang, Y.; Li, X. Role of ethylene crosstalk in seed germination and early seedling development: A review. Plant Physiol. Biochem. 2020, 151, 124–131.
- Iqbal, N.; Khan, N.A.; Ferrante, A.; Trivellini, A.; Francini, A.; Khan, M.I.R. Ethylene role in plant growth, development and senescence. Interaction with other phytohormones. Front. Plant Sci. 2017, 8, 475.
- Subbiah, V.; Reddy, K.J. Interactions between ethylene, abscisic acid and cytokinin during germination and seedling establishment in Arabidopsis. J. Biosci. 2010, 35, 451–458.
- Li, H.; Johnson, P.; Stepanova, A.; Alonso, J.M.; Ecker, J.R. Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev. Cell 2004, 7, 193–204.
- Zhong, S.; Zhao, M.; Shi, T.; Shi, H.; An, F.; Zhao, Q.; Guo, H. EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 2009, 106, 21431–21436.
- Dolgikh, V.A.; Pukhovaya, E.M.; Zemlyanskaya, E.V. Shaping ethylene response: The role of EIN3/EIL1 transcription factors. Front. Plant Sci. 2019, 10, 1030.
- Ceng-Hong, H.U.; Shi-Dong, Y.U.A.N.; Cui-Ling, T.O.N.G.; Zhang, D.J.; Huang, R.H. Ethylene modulates root growth and mineral nutrients levels in trifoliate orange through the auxin-signaling pathway. Not. Bot. Horti Agrobot. Cluj-Napoca 2023, 51, 13269.
- Wang, Z.; Yadav, V.; Yan, X.; Cheng, D.; Wei, C.; Zhang, X. Systematic genome-wide analysis of the ethylene-responsive ACS gene family: Contributions to sex form differentiation and development in melon and watermelon. Gene 2021, 805, 145910.
- Liu, Y.; Tang, M.; Liu, M.; Su, D.; Chen, J.; Gao, Y.; Li, Z. The molecular regulation of ethylene in fruit ripening. Small Methods 2020, 4, 1900485.
- Adams, D.O.; Yang, S. Ethylene biosynthesis: Identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Natl. Acad. Sci. USA 1979, 76, 170–174.
- Kende, H. Ethylene biosynthesis. Annu. Rev. Plant Biol. 1993, 44, 283–307.
- Houben, M.; Van de Poel, B. 1-Aminocyclopropane-1-carboxylic acid oxidase (ACO): The enzyme that makes the plant hormone ethylene. Front. Plant Sci. 2019, 10, 695.
- Qiao, H.; Chang, K.N.; Yazaki, J.; Ecker, J.R. Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 2009, 23, 512–521.
- Barry, C.S.; Llop-Tous, M.I.; Grierson, D. The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol. 2000, 123, 979–986.
- Ito, Y.; Kitagawa, M.; Ihashi, N.; Yabe, K.; Kimbara, J.; Yasuda, J.; Ito, H.; Inakuma, T.; Hiroi, S.; Kasumi, T. DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J. 2008, 55, 212–223.
- Zhang, Z.; Zhang, H.; Quan, R.; Wang, X.C.; Huang, R. Transcriptional regulation of the ethylene response factor LeERF2 in the expression of ethylene biosynthesis genes controls ethylene production in tomato and tobacco. Plant Physiol. 2009, 150, 365–377.
- Harkey, A.F.; Yoon, G.M.; Seo, D.H.; DeLong, A.; Muday, G.K. Light modulates ethylene synthesis, signaling, and downstream transcriptional networks to control plant development. Front. Plant Sci. 2019, 10, 1094.
- Pattyn, J.; Vaughan-Hirsch, J.; Van de Poel, B. The regulation of ethylene biosynthesis: A complex multilevel control circuitry. New Phytol. 2021, 229, 770–782.
- Tan, S.T.; Xue, H.W. Casein kinase 1 regulates ethylene synthesis by phosphorylating and promoting the turnover of ACS5. Cell Rep. 2014, 9, 1692–1702.
- Wen, X.; Zhang, C.; Ji, Y.; Zhao, Q.; He, W.; An, F.; Guo, H. Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 2012, 22, 1613–1616.
- Seo, D.H.; Yoon, G.M. Light-induced stabilization of ACS contributes to hypocotyl elongation during the dark-to-light transition in Arabidopsis seedlings. Plant J. 2019, 98, 898–911.
- Skottke, K.R.; Yoon, G.M.; Kieber, J.J.; DeLong, A. Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genet. 2011, 7, e1001370.
- Ruzicka, K.; Ljung, K.; Vanneste, S.; Podhorská, R.; Beeckman, T.; Friml, J.; Benková, E. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 2007, 19, 2197–2212.
- Ortega-Martínez, O.; Pernas, M.; Carol, R.J.; Dolan, L. Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 2007, 317, 507–510.
- Skirycz, A.; Claeys, H.; De Bodt, S.; Oikawa, A.; Shinoda, S.; Andriankaja, M.; Maleux, K.; Eloy, N.B.; Coppens, F.; Yoo, S.D.; et al. Pause-and-stop: The effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. Plant Cell 2011, 23, 1876–1888.
- Marsch-Martinez, N.; Greco, R.; Becker, J.D.; Dixit, S.; Bergervoet, J.H.; Karaba, A.; de Folter, S.; Pereira, A. BOLITA, an Arabidopsis AP2/ERF-like transcription factor that affects cell expansion and proliferation/differentiation pathways. Plant Mol. Biol. 2006, 62, 825–843.
- Dubois, M.; Van den Broeck, L.; Inzé, D. The pivotal role of ethylene in plant growth. Trends Plant Sci. 2018, 23, 311–323.
- Van de Poel, B.; Smet, D.; Van Der Straeten, D. Ethylene and hormonal cross talk in vegetative growth and development. Plant Physiol. 2015, 169, 61–72.
- Vogel, J.P.; Woeste, K.E.; Theologis, A.; Kieber, J.J. Recessive and dominant mutations in the ethylene biosynthetic gene ACS 5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc. Natl. Acad. Sci. USA 1998, 95, 4766–4771.
- Qu, X.; Hall, B.P.; Gao, Z.; Schaller, G.E. A strong constitutive ethylene-response phenotype conferred on Arabidopsis plants containing null mutations in the ethylene receptors ETR1 and ERS1. BMC Plant Biol. 2007, 7, 3.
- Feng, G.; Liu, G.; Xiao, J. The Arabidopsis EIN2 restricts organ growth by retarding cell expansion. Plant Signal. Behav. 2015, 10, e1017169.
- Ma, W.J.; Pannell, J.R. Sex determination: Separate sexes are a double turnoff in melons. Curr. Biol. 2016, 26, 171–174.
- Yin, T.; Quinn, J.A. A Mechanistic Model of a Single Hormone Regulating Both Sexes in Flowering Plants. Bull. Torrey Bot. Club 1992, 119, 431–441.
- Zhang, H.; Li, S.; Yang, L.; Cai, G.; Chen, H.; Gao, D.; Lin, T.; Cui, Q.; Wang, D.; Li, Z.; et al. Gain-of-function of the 1-aminocyclopropane-1-carboxylate synthase gene ACS1G induces female flower development in cucumber gynoecy. Plant Cell 2021, 33, 306–321.
- Chen, H.; Sun, J.; Li, S.; Cui, Q.; Zhang, H.; Xin, F.; Wang, H.; Lin, T.; Gao, D.; Wang, S.; et al. An ACC oxidase gene essential for cucumber carpel development. Mol. Plant 2016, 9, 1315–1327.
- Yu, D.; Li, X.; Li, Y.; Ali, F.; Li, F.; Wang, Z. Dynamic roles and intricate mechanisms of ethylene in epidermal hair development in Arabidopsis and cotton. New Phytol. 2022, 234, 375–391.
- Dolan, L. The role of ethylene in root hair growth in Arabidopsis. J. Plant. Nutr. Soil Sci. 2001, 164, 141–145.
- Liu, M.; Pirrello, J.; Chervin, C.; Roustan, J.P.; Bouzayen, M. Ethylene control of fruit ripening: Revisiting the complex network of transcriptional regulation. Plant Physiol. 2015, 169, 2380–2390.
- Nakatsuka, A.; Murachi, S.; Okunishi, H.; Shiomi, S.; Nakano, R.; Kubo, Y.; Inaba, A. Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant Physiol. 1998, 118, 1295–1305.
- Symons, G.M.; Chua, Y.J.; Ross, J.J.; Quittenden, L.J.; Davies, N.W.; Reid, J.B. Hormonal changes during non-climacteric ripening in strawberry. J. Exp. Bot. 2012, 63, 4741–4750.
- Cackett, L.; Luginbuehl, L.H.; Schreier, T.B.; Lopez-Juez, E.; Hibberd, J.M. Chloroplast development in green plant tissues: The interplay between light, hormone, and transcriptional regulation. New Phytol. 2022, 233, 2000–2016.
- Shi, H.; Liu, R.; Xue, C.; Shen, X.; Wei, N.; Deng, X.W.; Zhong, S. Seedlings transduce the depth and mechanical pressure of covering soil using COP1 and ethylene to regulate EBF1/EBF2 for soil emergence. Curr. Biol. 2016, 26, 139–149.
- An, F.; Zhao, Q.; Ji, Y.; Li, W.; Jiang, Z.; Yu, X.; Zhang, C.; Han, Y.; He, W.; Liu, Y.; et al. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 2010, 22, 2384–2401.
- Hernández-Verdeja, T.; Vuorijoki, L.; Strand, A. Emerging from the darkness: Interplay between light and plastid signaling during chloroplast biogenesis. Physiol. Plant. 2020, 169, 397–406.
- Chen, L.; Dodd, I.C.; Davies, W.J.; Wilkinson, S. Ethylene limits abscisic acid- or soil drying-induced stomatal closure in aged wheat leaves. Plant Cell Environ. 2013, 36, 1850–1859.
- Desikan, R.; Last, K.; Harrett-Williams, R.; Tagliavia, C.; Harter, K.; Hooley, R.; Hancock, J.T.; Neill, S.J. Ethylene-induced stomatal closure in Arabidopsis occurs via AtrbohF-mediated hydrogen peroxide synthesis. Plant J. 2006, 47, 907–916.
- Tholen, D.; Voesenek, L.A.C.J.; Poorter, H. Ethylene insensitivity does not increase leaf area or relative growth rate in Arabidopsis, Nicotiana tabacum, and Petunia x hybrida. Plant Physiol. 2004, 134, 1803–1812.
- Tanaka, Y.; Sano, T.; Tamaoki, M.; Nakajima, N.; Kondo, N.; Hasezawa, S. Ethylene inhibits abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol. 2005, 138, 2337–2343.
- Iqbal, N.; Nazar, R.; Syeed, S.; Masood, A.; Khan, N.A. Exogenously-sourced ethylene increases stomatal conductance, photosynthesis, and growth under optimal and deficient nitrogen fertilization in mustard. J. Exp. Bot. 2011, 62, 4955–4963.
- Ceusters, J.; Van de Poel, B. Ethylene exerts species-specific and age-dependent control of photosynthesis. Plant Physiol. 2018, 176, 2601–2612.
- Tholen, D.; Pons, T.L.; Voesenek, L.A.C.J.; Poorter, H. Ethylene insensitivity results in down-regulation of Rubisco expression and photosynthetic capacity in tobacco. Plant Physiol. 2007, 144, 1305–1315.
- Tholen, D.; Pons, T.L.; Voesenek, L.A.C.J.; Poorter, H. The role of ethylene perception in the control of photosynthesis. Plant Signal. Behav. 2008, 3, 108–109.
- Grbic, V.; Bleecker, A.B. Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 1995, 8, 595–602.
- Chen, Z.; Gallie, D.R. Ethylene regulates energy-dependent non-photochemical quenching in Arabidopsis through repression of the xanthophyll cycle. PLoS ONE 2015, 10, e0144209.
- Kim, G.D.; Cho, Y.H.; Yoo, S.D. Phytohormone ethylene-responsive Arabidopsis organ growth under light is in the fine regulation of Photosystem II deficiency-inducible AKIN10 expression. Sci. Rep. 2017, 7, 2767.
- Xie, X.L.; Xia, X.J.; Kuang, S.; Zhang, X.L.; Yin, X.R.; Yu, J.Q.; Chen, K.S. A novel ethylene responsive factor CitERF13 plays a role in photosynthesis regulation. Plant Sci. 2017, 256, 112–119.
- Woo, H.R.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf senescence: Systems and dynamics aspects. Annu. Rev. Plant Biol. 2019, 70, 347–376.
- van der Graaff, E.; Schwacke, R.; Schneider, A.; Desimone, M.; Flugge, U.I.; Kunze, R. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 2006, 141, 776–792.
- Tsuchisaka, A.; Yu, G.; Jin, H.; Alonso, J.M.; Ecker, J.R.; Zhang, X.; Gao, S.; Theologis, A. A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics 2009, 183, 979–1003.
- Kim, J.H.; Woo, H.R.; Kim, J.; Lim, P.O.; Lee, I.C.; Choi, S.H.; Hwang, D.; Nam, H.G. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 2009, 323, 1053–1057.
- Qiu, K.; Li, Z.; Yang, Z.; Chen, J.; Wu, S.; Zhu, X.; Gao, S.; Gao, J.; Ren, G.; Kuai, B.; et al. EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet. 2015, 11, e1005399.
- Zhang, Y.; Zang, Y.; Chen, J.; Feng, S.; Zhang, Z.; Hu, Y.; Zhang, T. A truncated ETHYLENE INSENSITIVE3-like protein, GhLYI, regulates senescence in cotton. Plant Physiol. 2023, 193, 1177–1196.
- Patharkar, O.R.; Walker, J.C. Advances in abscission signaling. J. Exp. Bot. 2018, 69, 733–740.
- Meir, S.; Philosoph-Hadas, S.; Riov, J.; Tucker, M.L.; Patterson, S.E.; Roberts, J.A. Re-evaluation of the ethylene-dependent and-independent pathways in the regulation of floral and organ abscission. J. Exp. Bot. 2019, 70, 1461–1467.
- Brown, K.M. Ethylene and abscission. Physiol. Plant. 1997, 100, 567–576.
- Botton, A.; Ruperti, B. The yes and no of the ethylene involvement in abscission. Plants 2019, 8, 187.
- Cheng, C.; Zhang, L.; Yang, X.; Zhong, G. Profiling gene expression in citrus fruit calyx abscission zone (AZ-C) treated with ethylene. Mol. Genet. Genom. 2015, 290, 1991–2006.
- Hartmond, U.; Yuan, R.; Burns, J.K.; Grant, A.; Kender, W.J. Citrus fruit abscission induced by methyl-jasmonate. J. Am. Soc. Hortic. Sci. 2000, 125, 547–552.
- Merelo, P.; Agustí, J.; Ventimilla, D.; Talón, M.; Tadeo, F.R. Vesicular trafficking in abscission zone cells during ethylene-promoted fruit abscission in citrus. Acta Hortic. 2019, 1230, 41–48.
- Dal Cin, V.; Barbaro, E.; Danesin, M.; Murayama, H.; Velasco, R.; Ramina, A. Fruitlet abscission: A cDNA-AFLP approach to study genes differentially expressed during shedding of immature fruits reveals the involvement of a putative auxin hydrogen symporter in apple (Malus domestica L. Borkh). Gene 2009, 442, 26–36.
- Li, J.; Zhu, H.; Yuan, R. Profiling the expression of genes related to ethylene biosynthesis, ethylene perception, and cell wall degradation during fruit abscission and fruit ripening in apple. J. Am. Soc. Hortic. Sci. 2010, 135, 391–401.
- Llop-Tous, I.; Barry, C.S.; Grierson, D. Regulation of ethylene biosynthesis in response to pollination in tomato flowers. Plant Physiol. 2000, 123, 971–978.
- Woltering, E.J.; van Doorn, W.C. Role of ethylene in senescence of petals: Morphological and taxonomical relationships. J. Exp. Bot. 1998, 208, 1605–1616.
More
Information
Subjects:
Plant Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
875
Revisions:
2 times
(View History)
Update Date:
19 Jan 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




