
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Spyros Skandalis | -- | 3251 | 2024-01-17 11:34:08 | | | |
| 2 | Lindsay Dong | Meta information modification | 3251 | 2024-01-19 02:56:31 | | | | |
| 3 | Lindsay Dong | Meta information modification | 3251 | 2024-01-22 02:54:51 | | |
Video Upload Options
CD44 serves as a cell surface receptor for various extracellular matrix molecules, mainly hyaluronan, and messenger molecules, such as growth factors, and has important functions in normal and disease states, the predominant one being cancer. CD44 coordinates both structural and signaling events through its highly conserved intracellular domain. Although short and devoid of any enzymatic activity, the CD44 intracellular domain possesses structural motifs that promote the interactions with cytoplasmic effectors involved in important cellular pathways, including cell trafficking, transcription, and metabolism, which regulate cellular functions like growth, survival, differentiation, stemness, and therapeutic resistance.
1. Introduction
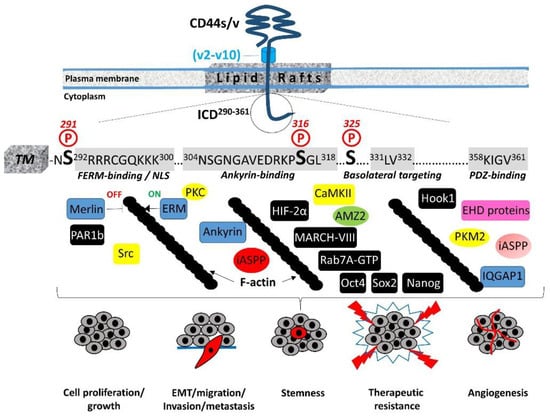
2. Structural Features of CD44 ICD
The transmembrane and intracellular domains of CD44 are required for its proper membrane localization, ligand binding, and functions such as cell adhesion and migration [4]. For instance, mutation or deletion of the ICD results in the aberrant localization of CD44 within cellular membranes, an inability to bind HA, and impaired HA-mediated cell migration and tumor development [13][14][15][16]. Therefore, these highly conserved domains are essential for ligand binding and subsequent downstream intracellular events (i.e., outside-in signaling), probably by promoting the stabilization and clustering of CD44 receptors at the plasma membrane [4][13][17]. In contrast, their precise amino acid sequence is not a prerequisite for CD44 functions, since the replacement of either domain with equivalent domains from different adhesion receptors does not impair binding to HA, cell adhesion to HA matrices, or rolling interaction of lymphoid cells with HA pericellular coats [13][18], suggesting that the shape and conformation of these domains do not affect the inside-out signaling of CD44.
3. Hyaluronan Internalization and Interactions with the Cell-Trafficking Machinery
4. Regulation of Cytoskeletal Organization and Cell Phenotype
5. Regulation of Cell–Cell Contact Inhibition and Cell Growth
6. Cleavage and Intracellular Release of CD44 ICD: A Master Regulator of the Transcriptome
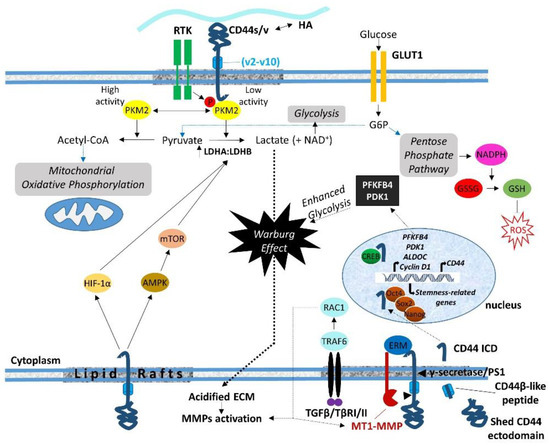
7. Regulation of Cell Metabolism
8. Conclusions
In conclusion, the association of CD44 ICD with cytoskeletal effectors (ERM, merlin, IQGAP1, and ankyrin) drives cytoskeleton rearrangements and affects the distribution of organelles and transport of molecules. In addition, through specific interactions of its cytoplasmic tail, CD44 regulates the cell-trafficking machinery, the transcriptome, and major cell metabolic pathways, with substantial impact on cell functional properties like survival, proliferation, adhesion, differentiation, therapeutic resistance, stemness properties, and EMT .
References
- Screaton, G.R.; Bell, M.V.; Jackson, D.G.; Cornelis, F.B.; Gerth, U.; Bell, J.I. Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc. Natl. Acad. Sci. USA 1992, 89, 12160–12164.
- Goldstein, L.A.; Zhou, D.F.; Picker, L.J.; Minty, C.N.; Bargatze, R.F.; Ding, J.F.; Butcher, E.C. A human lymphocyte homing receptor, the hermes antigen, is related to cartilage proteoglycan core and link proteins. Cell 1989, 56, 1063–1072.
- Isacke, C.M. The role of the cytoplasmic domain in regulating CD44 function. J. Cell Sci. 1994, 107 Pt 9, 2353–2359.
- Thorne, R.F.; Legg, J.W.; Isacke, C.M. The role of the CD44 transmembrane and cytoplasmic domains in co-ordinating adhesive and signalling events. J. Cell Sci. 2004, 117, 373–380.
- Heldin, P.; Kolliopoulos, C.; Lin, C.Y.; Heldin, C.H. Involvement of hyaluronan and CD44 in cancer and viral infections. Cell Signal. 2020, 65, 109427.
- Kincade, P.W.; Zheng, Z.; Katoh, S.; Hanson, L. The importance of cellular environment to function of the CD44 matrix receptor. Curr. Opin. Cell Biol. 1997, 9, 635–642.
- Morath, I.; Hartmann, T.N.; Orian-Rousseau, V. CD44: More than a mere stem cell marker. Int. J. Biochem. Cell Biol. 2016, 81, 166–173.
- Elkashty, O.A.; Elghanam, G.A.; Su, X.; Liu, Y.; Chauvin, P.J.; Tran, S.D. Cancer stem cells enrichment with surface markers CD271 and CD44 in human head and neck squamous cell carcinomas. Carcinogenesis 2020, 41, 458–466.
- Tsunekuni, K.; Konno, M.; Haraguchi, N.; Koseki, J.; Asai, A.; Matsuoka, K.; Kobunai, T.; Takechi, T.; Doki, Y.; Mori, M.; et al. CD44/CD133-Positive Colorectal Cancer Stem Cells are Sensitive to Trifluridine Exposure. Sci. Rep. 2019, 9, 14861.
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715.
- Li, J.; Zhou, B.P. Activation of beta-catenin and Akt pathways by Twist are critical for the maintenance of EMT associated cancer stem cell-like characters. BMC Cancer 2011, 11, 49.
- Skandalis, S.S.; Karalis, T.T.; Chatzopoulos, A.; Karamanos, N.K. Hyaluronan-CD44 axis orchestrates cancer stem cell functions. Cell Signal. 2019, 63, 109377.
- Lesley, J.; He, Q.; Miyake, K.; Hamann, A.; Hyman, R.; Kincade, P.W. Requirements for hyaluronic acid binding by CD44: A role for the cytoplasmic domain and activation by antibody. J. Exp. Med. 1992, 175, 257–266.
- Neame, S.J.; Isacke, C.M. The cytoplasmic tail of CD44 is required for basolateral localization in epithelial MDCK cells but does not mediate association with the detergent-insoluble cytoskeleton of fibroblasts. J. Cell Biol. 1993, 121, 1299–1310.
- Uff, C.R.; Neame, S.J.; Isacke, C.M. Hyaluronan binding by CD44 is regulated by a phosphorylation-independent mechanism. Eur. J. Immunol. 1995, 25, 1883–1887.
- Lokeshwar, V.B.; Fregien, N.; Bourguignon, L.Y. Ankyrin-binding domain of CD44(GP85) is required for the expression of hyaluronic acid-mediated adhesion function. J. Cell Biol. 1994, 126, 1099–1109.
- Thomas, L.; Byers, H.R.; Vink, J.; Stamenkovic, I. CD44H regulates tumor cell migration on hyaluronate-coated substrate. J. Cell Biol. 1992, 118, 971–977.
- Gal, I.; Lesley, J.; Ko, W.; Gonda, A.; Stoop, R.; Hyman, R.; Mikecz, K. Role of the extracellular and cytoplasmic domains of CD44 in the rolling interaction of lymphoid cells with hyaluronan under physiologic flow. J. Biol. Chem. 2003, 278, 11150–11158.
- Legg, J.W.; Isacke, C.M. Identification and functional analysis of the ezrin-binding site in the hyaluronan receptor, CD44. Curr Biol. 1998, 8, 705–708.
- Yonemura, S.; Hirao, M.; Doi, Y.; Takahashi, N.; Kondo, T.; Tsukita, S.; Tsukita, S. Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J. Cell Biol. 1998, 140, 885–895.
- Sheikh, H.; Isacke, C.M. A di-hydrophobic Leu-Val motif regulates the basolateral localization of CD44 in polarized Madin-Darby canine kidney epithelial cells. J. Biol. Chem. 1996, 271, 12185–12190.
- Heilker, R.; Spiess, M.; Crottet, P. Recognition of sorting signals by clathrin adaptors. Bioessays. 1999, 21, 558–567.
- Tammi, R.; Rilla, K.; Pienimaki, J.P.; MacCallum, D.K.; Hogg, M.; Luukkonen, M.; Hascall, V.C.; Tammi, M. Hyaluronan enters keratinocytes by a novel endocytic route for catabolism. J. Biol. Chem. 2001, 276, 35111–35122.
- Knudson, W.; Chow, G.; Knudson, C.B. CD44-mediated uptake and degradation of hyaluronan. Matrix Biol. 2002, 21, 15–23.
- Thankamony, S.P.; Knudson, W. Acylation of CD44 and its association with lipid rafts are required for receptor and hyaluronan endocytosis. J. Biol. Chem. 2006, 281, 34601–34609.
- Aguiar, D.J.; Knudson, W.; Knudson, C.B. Internalization of the hyaluronan receptor CD44 by chondrocytes. Exp. Cell Res. 1999, 252, 292–302.
- Eyster, C.A.; Cole, N.B.; Petersen, S.; Viswanathan, K.; Fruh, K.; Donaldson, J.G. MARCH ubiquitin ligases alter the itinerary of clathrin-independent cargo from recycling to degradation. Mol. Biol. Cell 2011, 22, 3218–3230.
- Skandalis, S.S.; Karalis, T.; Heldin, P. Intracellular hyaluronan: Importance for cellular functions. Semin Cancer Biol. 2020, 62, 20–30.
- Maldonado-Baez, L.; Donaldson, J.G. Hook1, microtubules, and Rab22: Mediators of selective sorting of clathrin-independent endocytic cargo proteins on endosomes. Bioarchitecture 2013, 3, 141–146.
- Skandalis, S.S.; Kozlova, I.; Engstrom, U.; Hellman, U.; Heldin, P. Proteomic identification of CD44 interacting proteins. IUBMB Life 2010, 62, 833–840.
- Murai, T. Lipid Raft-Mediated Regulation of Hyaluronan-CD44 Interactions in Inflammation and Cancer. Front. Immunol. 2015, 6, 420.
- Heiska, L.; Alfthan, K.; Gronholm, M.; Vilja, P.; Vaheri, A.; Carpen, O. Association of ezrin with intercellular adhesion molecule-1 and -2 (ICAM-1 and ICAM-2). Regulation by phosphatidylinositol 4, 5-bisphosphate. J. Biol. Chem. 1998, 273, 21893–21900.
- Granes, F.; Urena, J.M.; Rocamora, N.; Vilaro, S. Ezrin links syndecan-2 to the cytoskeleton. J. Cell Sci. 2000, 113 Pt 7, 1267–1276.
- Ivetic, A.; Deka, J.; Ridley, A.; Ager, A. The cytoplasmic tail of L-selectin interacts with members of the Ezrin-Radixin-Moesin (ERM) family of proteins: Cell activation-dependent binding of Moesin but not Ezrin. J. Biol. Chem. 2002, 277, 2321–2329.
- Obremski, V.J.; Hall, A.M.; Fernandez-Valle, C. Merlin, the neurofibromatosis type 2 gene product, and beta1 integrin associate in isolated and differentiating Schwann cells. J Neurobiol. 1998, 37, 487–501.
- Curto, M.; McClatchey, A.I. Nf2/Merlin: A coordinator of receptor signalling and intercellular contact. Br. J. Cancer 2008, 98, 256–262.
- Sato, T.; Sekido, Y. NF2/Merlin Inactivation and Potential Therapeutic Targets in Mesothelioma. Int. J. Mol. Sci. 2018, 19, 988.
- Mranda, G.M.; Xiang, Z.P.; Liu, J.J.; Wei, T.; Ding, Y. Advances in prognostic and therapeutic targets for hepatocellular carcinoma and intrahepatic cholangiocarcinoma: The hippo signaling pathway. Front. Oncol. 2022, 12, 937957.
- Ren, M.; Zhao, L.; Ma, Z.; An, H.; Marrink, S.J.; Sun, F. Molecular basis of PIP2-dependent conformational switching of phosphorylated CD44 in binding FERM. Biophys. J. 2023, 122, 2675–2685.
- Legg, J.W.; Lewis, C.A.; Parsons, M.; Ng, T.; Isacke, C.M. A novel PKC-regulated mechanism controls CD44 ezrin association and directional cell motility. Nat. Cell Biol. 2002, 4, 399–407.
- Bretscher, A.; Edwards, K.; Fehon, R.G. ERM proteins and merlin: Integrators at the cell cortex. Nat. Rev. Mol. Cell Biol. 2002, 3, 586–599.
- Ng, T.; Parsons, M.; Hughes, W.E.; Monypenny, J.; Zicha, D.; Gautreau, A.; Arpin, M.; Gschmeissner, S.; Verveer, P.J.; Bastiaens, P.I.; et al. Ezrin is a downstream effector of trafficking PKC-integrin complexes involved in the control of cell motility. EMBO J. 2001, 20, 2723–2741.
- Choi, Y.; Yun, J.H.; Yoo, J.; Lee, I.; Kim, H.; Son, H.N.; Kim, I.S.; Yoon, H.S.; Zimmermann, P.; Couchman, J.R.; et al. New structural insight of C-terminal region of Syntenin-1, enhancing the molecular dimerization and inhibitory function related on Syndecan-4 signaling. Sci. Rep. 2016, 6, 36818.
- Iuliano, R.; Trapasso, F.; Sama, I.; Le Pera, I.; Martelli, M.L.; Lembo, F.; Santoro, M.; Viglietto, G.; Chiariotti, L.; Fusco, A. Rat protein tyrosine phosphatase eta physically interacts with the PDZ domains of syntenin. FEBS Lett. 2001, 500, 41–44.
- Bass, M.D.; Humphries, M.J. Cytoplasmic interactions of syndecan-4 orchestrate adhesion receptor and growth factor receptor signalling. Biochem. J. 2002, 368, 1–15.
- Bourguignon, L.Y.; Zhu, H.; Shao, L.; Zhu, D.; Chen, Y.W. Rho-kinase (ROK) promotes CD44v(3,8-10)-ankyrin interaction and tumor cell migration in metastatic breast cancer cells. Cell Motil. Cytoskelet. 1999, 43, 269–287.
- Oliferenko, S.; Paiha, K.; Harder, T.; Gerke, V.; Schwarzler, C.; Schwarz, H.; Beug, H.; Gunthert, U.; Huber, L.A. Analysis of CD44-containing lipid rafts: Recruitment of annexin II and stabilization by the actin cytoskeleton. J. Cell Biol. 1999, 146, 843–854.
- Bourguignon, L.Y.; Singleton, P.A.; Diedrich, F.; Stern, R.; Gilad, E. CD44 interaction with Na+-H+ exchanger (NHE1) creates acidic microenvironments leading to hyaluronidase-2 and cathepsin B activation and breast tumor cell invasion. J. Biol. Chem. 2004, 279, 26991–27007.
- Triantafilou, M.; Miyake, K.; Golenbock, D.T.; Triantafilou, K. Mediators of innate immune recognition of bacteria concentrate in lipid rafts and facilitate lipopolysaccharide-induced cell activation. J. Cell Sci. 2002, 115, 2603–2611.
- Grass, G.D.; Dai, L.; Qin, Z.; Parsons, C.; Toole, B.P. CD147: Regulator of hyaluronan signaling in invasiveness and chemoresistance. Adv. Cancer Res. 2014, 123, 351–373.
- Wu, A.; Wojtowicz, K.; Savary, S.; Hamon, Y.; Trombik, T. Do ABC transporters regulate plasma membrane organization? Cell. Mol. Biol. Lett. 2020, 25, 37.
- Xu, Y.; Yu, Q. E-cadherin negatively regulates CD44-hyaluronan interaction and CD44-mediated tumor invasion and branching morphogenesis. J. Biol. Chem. 2003, 278, 8661–8668.
- Guo, Y.J.; Lin, S.C.; Wang, J.H.; Bigby, M.; Sy, M.S. Palmitoylation of CD44 interferes with CD3-mediated signaling in human T lymphocytes. Int. Immunol. 1994, 6, 213–221.
- Freeman, S.A.; Vega, A.; Riedl, M.; Collins, R.F.; Ostrowski, P.P.; Woods, E.C.; Bertozzi, C.R.; Tammi, M.I.; Lidke, D.S.; Johnson, P.; et al. Transmembrane Pickets Connect Cyto- and Pericellular Skeletons Forming Barriers to Receptor Engagement. Cell 2018, 172, 305–317.e10.
- Hu, S.; Shi, X.; Liu, Y.; He, Y.; Du, Y.; Zhang, G.; Yang, C.; Gao, F. CD44 cross-linking increases malignancy of breast cancer via upregulation of p-Moesin. Cancer Cell Int. 2020, 20, 563.
- Brown, K.L.; Birkenhead, D.; Lai, J.C.; Li, L.; Li, R.; Johnson, P. Regulation of hyaluronan binding by F-actin and colocalization of CD44 and phosphorylated ezrin/radixin/moesin (ERM) proteins in myeloid cells. Exp. Cell Res. 2005, 303, 400–414.
- Liu, D.; Liu, T.; Sy, M.S. Identification of two regions in the cytoplasmic domain of CD44 through which PMA, calcium, and foskolin differentially regulate the binding of CD44 to hyaluronic acid. Cell Immunol. 1998, 190, 132–140.
- Seluanov, A.; Hine, C.; Azpurua, J.; Feigenson, M.; Bozzella, M.; Mao, Z.; Catania, K.C.; Gorbunova, V. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc. Natl. Acad. Sci. USA 2009, 106, 19352–19357.
- Tian, X.; Azpurua, J.; Hine, C.; Vaidya, A.; Myakishev-Rempel, M.; Ablaeva, J.; Mao, Z.; Nevo, E.; Gorbunova, V.; Seluanov, A. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 2013, 499, 346–349.
- Morrison, H.; Sherman, L.S.; Legg, J.; Banine, F.; Isacke, C.; Haipek, C.A.; Gutmann, D.H.; Ponta, H.; Herrlich, P. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 2001, 15, 968–980.
- Jin, H.; Sperka, T.; Herrlich, P.; Morrison, H. Tumorigenic transformation by CPI-17 through inhibition of a merlin phosphatase. Nature 2006, 442, 576–579.
- Hartmann, M.; Parra, L.M.; Ruschel, A.; Bohme, S.; Li, Y.; Morrison, H.; Herrlich, A.; Herrlich, P. Tumor Suppressor NF2 Blocks Cellular Migration by Inhibiting Ectodomain Cleavage of CD44. Mol. Cancer Res. 2015, 13, 879–890.
- Ooki, T.; Murata-Kamiya, N.; Takahashi-Kanemitsu, A.; Wu, W.; Hatakeyama, M. High-Molecular-Weight Hyaluronan Is a Hippo Pathway Ligand Directing Cell Density-Dependent Growth Inhibition via PAR1b. Dev. Cell 2019, 49, 590–604.e9.
- Lichtenthaler, S.F.; Lemberg, M.K.; Fluhrer, R. Proteolytic ectodomain shedding of membrane proteins in mammals-hardware, concepts, and recent developments. EMBO J. 2018, 37, e99456.
- Hartmann, M.; Parra, L.M.; Ruschel, A.; Lindner, C.; Morrison, H.; Herrlich, A.; Herrlich, P. Inside-out Regulation of Ectodomain Cleavage of Cluster-of-Differentiation-44 (CD44) and of Neuregulin-1 Requires Substrate Dimerization. J. Biol. Chem. 2015, 290, 17041–17054.
- Wöhner, B.; Li, W.; Hey, S.; Drobny, A.; Werny, L.; Becker-Pauly, C.; Lucius, R.; Zunke, F.; Linder, S.; Arnold, P. Proteolysis of CD44 at the cell surface controls a downstream protease network. Front. Mol. Biosci. 2023, 10, 1026810.
- Suenaga, N.; Mori, H.; Itoh, Y.; Seiki, M. CD44 binding through the hemopexin-like domain is critical for its shedding by membrane-type 1 matrix metalloproteinase. Oncogene 2005, 24, 859–868.
- Terawaki, S.; Kitano, K.; Aoyama, M.; Mori, T.; Hakoshima, T. MT1-MMP recognition by ERM proteins and its implication in CD44 shedding. Genes Cells 2015, 20, 847–859.
- Tamada, M.; Nagano, O.; Tateyama, S.; Ohmura, M.; Yae, T.; Ishimoto, T.; Sugihara, E.; Onishi, N.; Yamamoto, T.; Yanagawa, H.; et al. Modulation of glucose metabolism by CD44 contributes to antioxidant status and drug resistance in cancer cells. Cancer Res. 2012, 72, 1438–1448.
- Christofk, H.R.; Vander Heiden, M.G.; Harris, M.H.; Ramanathan, A.; Gerszten, R.E.; Wei, R.; Fleming, M.D.; Schreiber, S.L.; Cantley, L.C. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 2008, 452, 230–233.
- Christofk, H.R.; Vander Heiden, M.G.; Wu, N.; Asara, J.M.; Cantley, L.C. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 2008, 452, 181–186.
- Hitosugi, T.; Kang, S.; Vander Heiden, M.G.; Chung, T.W.; Elf, S.; Lythgoe, K.; Dong, S.; Lonial, S.; Wang, X.; Chen, G.Z.; et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci. Signal. 2009, 2, ra73.
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033.
- Matoba, S.; Kang, J.G.; Patino, W.D.; Wragg, A.; Boehm, M.; Gavrilova, O.; Hurley, P.J.; Bunz, F.; Hwang, P.M. p53 regulates mitochondrial respiration. Science 2006, 312, 1650–1653.
- Miletti-Gonzalez, K.E.; Murphy, K.; Kumaran, M.N.; Ravindranath, A.K.; Wernyj, R.P.; Kaur, S.; Miles, G.D.; Lim, E.; Chan, R.; Chekmareva, M.; et al. Identification of function for CD44 intracytoplasmic domain (CD44-ICD): Modulation of matrix metalloproteinase 9 (MMP-9) transcription via novel promoter response element. J. Biol. Chem. 2012, 287, 18995–19007.
- Gao, R.; Li, D.; Xun, J.; Zhou, W.; Li, J.; Wang, J.; Liu, C.; Li, X.; Shen, W.; Qiao, H.; et al. CD44ICD promotes breast cancer stemness via PFKFB4-mediated glucose metabolism. Theranostics 2018, 8, 6248–6262.





