CD44 serves as a cell surface receptor for various extracellular matrix molecules, mainly hyaluronan, and messenger molecules, such as growth factors, and has important functions in normal and disease states, the predominant one being cancer. CD44 coordinates both structural and signaling events through its highly conserved intracellular domain. Although short and devoid of any enzymatic activity, the CD44 intracellular domain possesses structural motifs that promote the interactions with cytoplasmic effectors involved in important cellular pathways, including cell trafficking, transcription, and metabolism, which regulate cellular functions like growth, survival, differentiation, stemness, and therapeutic resistance.
- CD44
- cancer
- transcriptome
- cell metabolism
- cytoskeleton
- cell trafficking
- contact inhibition
1. Introduction
2. Structural Features of CD44 ICD
The transmembrane and intracellular domains of CD44 are required for its proper membrane localization, ligand binding, and functions such as cell adhesion and migration [4]. For instance, mutation or deletion of the ICD results in the aberrant localization of CD44 within cellular membranes, an inability to bind HA, and impaired HA-mediated cell migration and tumor development [13][14][15][16]. Therefore, these highly conserved domains are essential for ligand binding and subsequent downstream intracellular events (i.e., outside-in signaling), probably by promoting the stabilization and clustering of CD44 receptors at the plasma membrane [4][13][17]. In contrast, their precise amino acid sequence is not a prerequisite for CD44 functions, since the replacement of either domain with equivalent domains from different adhesion receptors does not impair binding to HA, cell adhesion to HA matrices, or rolling interaction of lymphoid cells with HA pericellular coats [13][18], suggesting that the shape and conformation of these domains do not affect the inside-out signaling of CD44.
The 72-amino-acid-residue cytoplasmic tail contains two positively charged amino acid clusters in the juxtamembrane domain (292RRRCGQKKK300), which constitute the FERM-binding domain that mediates the interaction of CD44 with ERM (ezrin/radixin/moesin) cytoskeletal proteins [19][20]. This sequence also contains the putative acylation site Cys295, suggesting that partition of CD44 into lipid rafts may regulate CD44 association with ERM proteins (see Section 4, “Regulation of cytoskeletal organization and cell phenotype”). The FERM-binding domain is followed by the ankyrin-binding domain (304NSGNGAVEDRKPSGL318) [16], an additional cytoskeleton association site, the dihydrophobic basolateral targeting motif 331LV332 [21], and four C-terminal amino acids (358KIGV361) that represent the PDZ (PSD-95/Dlg/ZO-1)-domain-binding peptide [4] (Figure 1).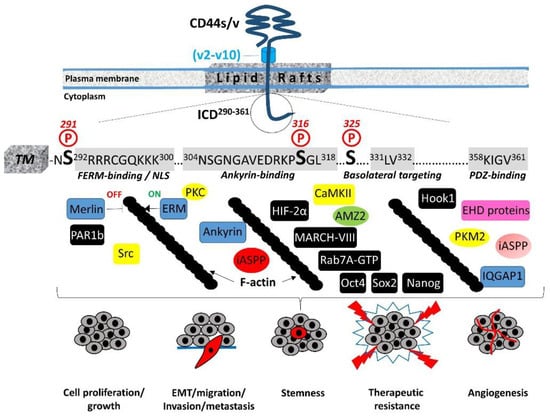
3. Hyaluronan Internalization and Interactions with the Cell-Trafficking Machinery
4. Regulation of Cytoskeletal Organization and Cell Phenotype
In migrating cells, CD44 shows a preferential distribution in actin polymerization regions such as the lamellipodia, filopodia, and apical microvilli, suggesting a role of CD44 in the regulation of actin cytoskeleton reorganization. This regulation is not direct, since CD44 ICD does not contain any binding sites for actin filaments but instead structural motifs such as the FERM (4.1 protein, Ezrin, Radixin, Moesin)-binding domain and the ankyrin-binding domain, which allow CD44 to connect and interact with the cytoskeleton. Specific clusters of basic residues (292RRRCGQKKK300) in the cytoplasmic tail of CD44 constitute the FERM-binding domain that interacts with ERM proteins and merlin/NF2 followed by a structural motif containing Ser316 (304NSGNGAVEDRKPSGL318) that binds the ankyrin cytoskeletal protein [4] (Figure 1). The ERM proteins and merlin are binding partners for a number of transmembrane receptors (like ICAM, syndecans, L-selectin, and integrins) acting as cross-linkers between the plasma membrane and the cortical actin filaments [32][33][34][35]. The neurofibromatosis type 2 (NF2) gene encodes the merlin protein, which has tumor-suppressing functions through regulating Hippo signaling, as well as receptor tyrosine kinases and downstream signal transduction pathways [36][37][38]. ERM proteins and merlin show a similar domain organization, with the highest homology in the conserved three-lobe N-terminal FERM domain (head). Phosphorylation of ERM proteins promotes their transition from the closed “inactive” conformation (head-to-tail self-association) to the open “active” conformation (dissociation of the three-lobed structure from the C-terminal domain) and their recruitment to the plasma membrane, where they bind membrane phospholipids such as phosphatidylinositol 4,5-biphosphate (PIP2). These interactions stabilize ERM proteins close to membrane-associated molecules such as CD44, which adopts a more open conformation at its ICD upon Ser325 phosphorylation [39]. Activated ERM binds to the FERM-binding domain of CD44 ICD phosphorylated at Ser325 (a constitutive phosphorylation site operated by CaMKII), forming a dynamic CD44-cytoskeleton association. This association can be disrupted upon PKC activation, which triggers the complete dephosphorylation of Ser325 followed by the phosphorylation of other Ser residues such as Ser291 and Ser316, leading to dissociation of the ERM proteins from CD44 and its disengagement from the actin cytoskeleton [4][40][41]. Therefore, the phosphorylation status of CD44 at specific Ser residues within the ICD is crucial for the dynamic association of the receptor with the cytoskeletal network and CD44-mediated chemotaxis, motility, and cell phenotypic changes. These events are not random but instead fine-tuned by the coordinated action of kinases and phosphatases. These enzymes could bind directly or indirectly to CD44. For example, ezrin interacts with PKC, thereby acting as a scaffolding protein for other signaling molecules to regulate phosphorylation of CD44 and associated proteins/receptors [42]. On the other hand, the existence of a PDZ-binding motif in CD44 ICD raises the possibility of a direct interaction of CD44 with PDZ domain-containing phosphatases or indirectly through PDZ domain adaptor proteins, which in turn bind protein phosphatases, thereby regulating the phosphorylation level of CD44 itself as well as other adjacent signaling molecules/receptors. For example, binding of syntenin, a syndecan-binding PDZ protein [43], to the C-terminus of protein tyrosine phosphatase (PTP) eta could result in the recruitment of this transmembrane PTP into syndecan-containing complexes [44][45]. The interaction of CD44 with the actin cytoskeleton through binding to ERM proteins and ankyrin is further stabilized by the partitioning of CD44 into detergent-insoluble, cholesterol-rich nano-domains (lipid rafts) of the plasma membrane [4][46][47] (Figure 1). The highly conserved intramembrane domain of CD44 can be reversibly palmitoylated at Cys286 and Cys295, which enhances localization of the receptor in cellular membranes as well as its interactions with cell surface and cytoplasmic proteins, such as several RTKs, innate receptors (TLRs), ABC transporters, EMMPRIN, PI3K, Src kinases, ezrin, and hyaluronidase 2, which preferentially localize into lipid rafts [48][49][50][51]. Interestingly, the amount of CD44 in lipid rafts can vary in different cell types and can be displaced from these membrane domains upon E-cadherin expression, which negatively regulates HA–CD44 interactions and CD44-dependent tumor invasion and branching morphogenesis [52]. In immune cells, CD44 is directed into lipid rafts via palmitoylation, which impairs the CD3-mediated signaling [53]. The strong association between CD44 and the actin filament network causes CD44 to act like a transmembrane picket that connects the cytoskeleton to pericellular milieu, forming barriers against diffusion of molecules through the plasma membrane [54]. Of particular interest is the cross-linking of CD44 receptors, which has been associated with the metastatic potential of several human cancers. Receptor aggregation can be drastically induced by both extracellular (pericellular HMW HA) and intracellular (phosphorylated moesin/ezrin) cues [55]. The association of CD44 with ezrin and actin promotes HA binding to cells [56][57].5. Regulation of Cell–Cell Contact Inhibition and Cell Growth
CD44-mediated contact inhibition of cell growth is modulated by counteracting CD44 ICD interacting proteins; e.g., the tumor suppressor protein merlin and ERM proteins. Their reversible dynamic association with CD44 ICD regulates the effects of the receptor on cell growth and motility since it provides an ON (CD44–ERM)/OFF (CD44–merlin) mechanism in the CD44–actin cytoskeleton association and signal transduction pathways (Figure 1). The importance of these interactions in cell physiology is evidenced by the finding that long-lived naked mole rats, which synthesize HA of exceptionally high molecular size, show a remarkable resistance to cancer through regulation of cell–cell contact inhibition via the HA–CD44–merlin signaling axis [58][59][60]. Mechanistically, ERM proteins and merlin are both hypophosphorylated in high-cell-density conditions [61]. Under these conditions, ERM proteins are inactive, while merlin adopts the active unphosphorylated closed conformation and interacts with HA-bound CD44 and E-cadherin, thus stabilizing the homophilic E-cadherin interactions and cell–cell adhesion complexes and rendering the cells stationary. Activated merlin prevents proteolytic processing of the CD44 extracellular domain, which in turn preserves cell density signaling and inhibits cell proliferation and migration [62]. In addition to merlin, PAR1b (partitioning defective 1b) also links CD44 and the Hippo pathway [63]. Ooki and colleagues found that HMW–HA-mediated CD44 clustering induces the interaction of CD44 ICD with the PAR1b/microtubule affinity-regulating kinase 2 (MARK2) complex, which normally inactivates MST1 and MST2 Ser/Thr kinases through binding and subsequent inhibitory phosphorylation, thus triggering Hippo signaling activation and contributing to contact inhibition of cell growth.6. Cleavage and Intracellular Release of CD44 ICD: A Master Regulator of the Transcriptome
The proteolytic cleavage of the extracellular domain or shedding of membrane-associated proteins is an irreversible post-translational modification that regulates cell–cell communication, intercellular signaling, and biological functions by releasing growth factors, enzymes, and soluble receptors [64]. In some instances, the remaining protein in the membrane is further cleaved, generating additional products that include a cytoplasmic fragment that can function in intracellular signaling, revealing an additional level of regulation of cellular functions. In line with this, CD44 can undergo successive proteolytic processing by a number of proteases, resulting in the generation of both extra- and intracellular bioactive fragments. These proteases are recruited and act at the cleavage site under specific prerequisites. The formation of CD44 dimers has been suggested to be critical [65]. The sequential cleavage of CD44 begins with the ectodomain shedding by transmembrane MMPs (i.e., MT1-MMP and MT3-MMP) as well as other CD44 sheddases (i.e., disintegrin or ADAM 10, ADAM 17, and meprin β), resulting in the generation of a membrane-associated fragment of CD44 while its extracellular N-terminal region is released to the ECM [5][66]. ERM proteins are involved in CD44 shedding, since they mediate the colocalization of MT1-MMP and CD44. In particular, the radixin FERM domain simultaneously binds the cytoplasmic tails of MT1-MMP and CD44, forming a complex that is stabilized through anchoring to filamentous actin. This ternary complex facilitates the recognition of the CD44 stem region by the PEX domain of the MT1-MMP ectodomain, resulting in CD44 shedding [67][68] (Figure 2).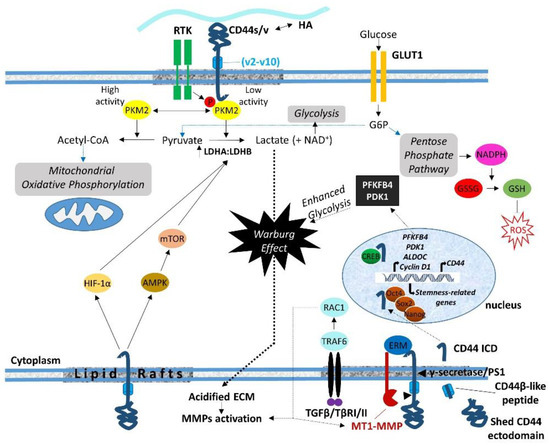
7. Regulation of Cell Metabolism
8. Conclusions
In conclusion, the association of CD44 ICD with cytoskeletal effectors (ERM, merlin, IQGAP1, and ankyrin) drives cytoskeleton rearrangements and affects the distribution of organelles and transport of molecules. In addition, through specific interactions of its cytoplasmic tail, CD44 regulates the cell-trafficking machinery, the transcriptome, and major cell metabolic pathways, with substantial impact on cell functional properties like survival, proliferation, adhesion, differentiation, therapeutic resistance, stemness properties, and EMT .
References
- Screaton, G.R.; Bell, M.V.; Jackson, D.G.; Cornelis, F.B.; Gerth, U.; Bell, J.I. Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc. Natl. Acad. Sci. USA 1992, 89, 12160–12164.
- Goldstein, L.A.; Zhou, D.F.; Picker, L.J.; Minty, C.N.; Bargatze, R.F.; Ding, J.F.; Butcher, E.C. A human lymphocyte homing receptor, the hermes antigen, is related to cartilage proteoglycan core and link proteins. Cell 1989, 56, 1063–1072.
- Isacke, C.M. The role of the cytoplasmic domain in regulating CD44 function. J. Cell Sci. 1994, 107 Pt 9, 2353–2359.
- Thorne, R.F.; Legg, J.W.; Isacke, C.M. The role of the CD44 transmembrane and cytoplasmic domains in co-ordinating adhesive and signalling events. J. Cell Sci. 2004, 117, 373–380.
- Heldin, P.; Kolliopoulos, C.; Lin, C.Y.; Heldin, C.H. Involvement of hyaluronan and CD44 in cancer and viral infections. Cell Signal. 2020, 65, 109427.
- Kincade, P.W.; Zheng, Z.; Katoh, S.; Hanson, L. The importance of cellular environment to function of the CD44 matrix receptor. Curr. Opin. Cell Biol. 1997, 9, 635–642.
- Morath, I.; Hartmann, T.N.; Orian-Rousseau, V. CD44: More than a mere stem cell marker. Int. J. Biochem. Cell Biol. 2016, 81, 166–173.
- Elkashty, O.A.; Elghanam, G.A.; Su, X.; Liu, Y.; Chauvin, P.J.; Tran, S.D. Cancer stem cells enrichment with surface markers CD271 and CD44 in human head and neck squamous cell carcinomas. Carcinogenesis 2020, 41, 458–466.
- Tsunekuni, K.; Konno, M.; Haraguchi, N.; Koseki, J.; Asai, A.; Matsuoka, K.; Kobunai, T.; Takechi, T.; Doki, Y.; Mori, M.; et al. CD44/CD133-Positive Colorectal Cancer Stem Cells are Sensitive to Trifluridine Exposure. Sci. Rep. 2019, 9, 14861.
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715.
- Li, J.; Zhou, B.P. Activation of beta-catenin and Akt pathways by Twist are critical for the maintenance of EMT associated cancer stem cell-like characters. BMC Cancer 2011, 11, 49.
- Skandalis, S.S.; Karalis, T.T.; Chatzopoulos, A.; Karamanos, N.K. Hyaluronan-CD44 axis orchestrates cancer stem cell functions. Cell Signal. 2019, 63, 109377.
- Lesley, J.; He, Q.; Miyake, K.; Hamann, A.; Hyman, R.; Kincade, P.W. Requirements for hyaluronic acid binding by CD44: A role for the cytoplasmic domain and activation by antibody. J. Exp. Med. 1992, 175, 257–266.
- Neame, S.J.; Isacke, C.M. The cytoplasmic tail of CD44 is required for basolateral localization in epithelial MDCK cells but does not mediate association with the detergent-insoluble cytoskeleton of fibroblasts. J. Cell Biol. 1993, 121, 1299–1310.
- Uff, C.R.; Neame, S.J.; Isacke, C.M. Hyaluronan binding by CD44 is regulated by a phosphorylation-independent mechanism. Eur. J. Immunol. 1995, 25, 1883–1887.
- Lokeshwar, V.B.; Fregien, N.; Bourguignon, L.Y. Ankyrin-binding domain of CD44(GP85) is required for the expression of hyaluronic acid-mediated adhesion function. J. Cell Biol. 1994, 126, 1099–1109.
- Thomas, L.; Byers, H.R.; Vink, J.; Stamenkovic, I. CD44H regulates tumor cell migration on hyaluronate-coated substrate. J. Cell Biol. 1992, 118, 971–977.
- Gal, I.; Lesley, J.; Ko, W.; Gonda, A.; Stoop, R.; Hyman, R.; Mikecz, K. Role of the extracellular and cytoplasmic domains of CD44 in the rolling interaction of lymphoid cells with hyaluronan under physiologic flow. J. Biol. Chem. 2003, 278, 11150–11158.
- Legg, J.W.; Isacke, C.M. Identification and functional analysis of the ezrin-binding site in the hyaluronan receptor, CD44. Curr Biol. 1998, 8, 705–708.
- Yonemura, S.; Hirao, M.; Doi, Y.; Takahashi, N.; Kondo, T.; Tsukita, S.; Tsukita, S. Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J. Cell Biol. 1998, 140, 885–895.
- Sheikh, H.; Isacke, C.M. A di-hydrophobic Leu-Val motif regulates the basolateral localization of CD44 in polarized Madin-Darby canine kidney epithelial cells. J. Biol. Chem. 1996, 271, 12185–12190.
- Heilker, R.; Spiess, M.; Crottet, P. Recognition of sorting signals by clathrin adaptors. Bioessays. 1999, 21, 558–567.
- Tammi, R.; Rilla, K.; Pienimaki, J.P.; MacCallum, D.K.; Hogg, M.; Luukkonen, M.; Hascall, V.C.; Tammi, M. Hyaluronan enters keratinocytes by a novel endocytic route for catabolism. J. Biol. Chem. 2001, 276, 35111–35122.
- Knudson, W.; Chow, G.; Knudson, C.B. CD44-mediated uptake and degradation of hyaluronan. Matrix Biol. 2002, 21, 15–23.
- Thankamony, S.P.; Knudson, W. Acylation of CD44 and its association with lipid rafts are required for receptor and hyaluronan endocytosis. J. Biol. Chem. 2006, 281, 34601–34609.
- Aguiar, D.J.; Knudson, W.; Knudson, C.B. Internalization of the hyaluronan receptor CD44 by chondrocytes. Exp. Cell Res. 1999, 252, 292–302.
- Eyster, C.A.; Cole, N.B.; Petersen, S.; Viswanathan, K.; Fruh, K.; Donaldson, J.G. MARCH ubiquitin ligases alter the itinerary of clathrin-independent cargo from recycling to degradation. Mol. Biol. Cell 2011, 22, 3218–3230.
- Skandalis, S.S.; Karalis, T.; Heldin, P. Intracellular hyaluronan: Importance for cellular functions. Semin Cancer Biol. 2020, 62, 20–30.
- Maldonado-Baez, L.; Donaldson, J.G. Hook1, microtubules, and Rab22: Mediators of selective sorting of clathrin-independent endocytic cargo proteins on endosomes. Bioarchitecture 2013, 3, 141–146.
- Skandalis, S.S.; Kozlova, I.; Engstrom, U.; Hellman, U.; Heldin, P. Proteomic identification of CD44 interacting proteins. IUBMB Life 2010, 62, 833–840.
- Murai, T. Lipid Raft-Mediated Regulation of Hyaluronan-CD44 Interactions in Inflammation and Cancer. Front. Immunol. 2015, 6, 420.
- Heiska, L.; Alfthan, K.; Gronholm, M.; Vilja, P.; Vaheri, A.; Carpen, O. Association of ezrin with intercellular adhesion molecule-1 and -2 (ICAM-1 and ICAM-2). Regulation by phosphatidylinositol 4, 5-bisphosphate. J. Biol. Chem. 1998, 273, 21893–21900.
- Granes, F.; Urena, J.M.; Rocamora, N.; Vilaro, S. Ezrin links syndecan-2 to the cytoskeleton. J. Cell Sci. 2000, 113 Pt 7, 1267–1276.
- Ivetic, A.; Deka, J.; Ridley, A.; Ager, A. The cytoplasmic tail of L-selectin interacts with members of the Ezrin-Radixin-Moesin (ERM) family of proteins: Cell activation-dependent binding of Moesin but not Ezrin. J. Biol. Chem. 2002, 277, 2321–2329.
- Obremski, V.J.; Hall, A.M.; Fernandez-Valle, C. Merlin, the neurofibromatosis type 2 gene product, and beta1 integrin associate in isolated and differentiating Schwann cells. J Neurobiol. 1998, 37, 487–501.
- Curto, M.; McClatchey, A.I. Nf2/Merlin: A coordinator of receptor signalling and intercellular contact. Br. J. Cancer 2008, 98, 256–262.
- Sato, T.; Sekido, Y. NF2/Merlin Inactivation and Potential Therapeutic Targets in Mesothelioma. Int. J. Mol. Sci. 2018, 19, 988.
- Mranda, G.M.; Xiang, Z.P.; Liu, J.J.; Wei, T.; Ding, Y. Advances in prognostic and therapeutic targets for hepatocellular carcinoma and intrahepatic cholangiocarcinoma: The hippo signaling pathway. Front. Oncol. 2022, 12, 937957.
- Ren, M.; Zhao, L.; Ma, Z.; An, H.; Marrink, S.J.; Sun, F. Molecular basis of PIP2-dependent conformational switching of phosphorylated CD44 in binding FERM. Biophys. J. 2023, 122, 2675–2685.
- Legg, J.W.; Lewis, C.A.; Parsons, M.; Ng, T.; Isacke, C.M. A novel PKC-regulated mechanism controls CD44 ezrin association and directional cell motility. Nat. Cell Biol. 2002, 4, 399–407.
- Bretscher, A.; Edwards, K.; Fehon, R.G. ERM proteins and merlin: Integrators at the cell cortex. Nat. Rev. Mol. Cell Biol. 2002, 3, 586–599.
- Ng, T.; Parsons, M.; Hughes, W.E.; Monypenny, J.; Zicha, D.; Gautreau, A.; Arpin, M.; Gschmeissner, S.; Verveer, P.J.; Bastiaens, P.I.; et al. Ezrin is a downstream effector of trafficking PKC-integrin complexes involved in the control of cell motility. EMBO J. 2001, 20, 2723–2741.
- Choi, Y.; Yun, J.H.; Yoo, J.; Lee, I.; Kim, H.; Son, H.N.; Kim, I.S.; Yoon, H.S.; Zimmermann, P.; Couchman, J.R.; et al. New structural insight of C-terminal region of Syntenin-1, enhancing the molecular dimerization and inhibitory function related on Syndecan-4 signaling. Sci. Rep. 2016, 6, 36818.
- Iuliano, R.; Trapasso, F.; Sama, I.; Le Pera, I.; Martelli, M.L.; Lembo, F.; Santoro, M.; Viglietto, G.; Chiariotti, L.; Fusco, A. Rat protein tyrosine phosphatase eta physically interacts with the PDZ domains of syntenin. FEBS Lett. 2001, 500, 41–44.
- Bass, M.D.; Humphries, M.J. Cytoplasmic interactions of syndecan-4 orchestrate adhesion receptor and growth factor receptor signalling. Biochem. J. 2002, 368, 1–15.
- Bourguignon, L.Y.; Zhu, H.; Shao, L.; Zhu, D.; Chen, Y.W. Rho-kinase (ROK) promotes CD44v(3,8-10)-ankyrin interaction and tumor cell migration in metastatic breast cancer cells. Cell Motil. Cytoskelet. 1999, 43, 269–287.
- Oliferenko, S.; Paiha, K.; Harder, T.; Gerke, V.; Schwarzler, C.; Schwarz, H.; Beug, H.; Gunthert, U.; Huber, L.A. Analysis of CD44-containing lipid rafts: Recruitment of annexin II and stabilization by the actin cytoskeleton. J. Cell Biol. 1999, 146, 843–854.
- Bourguignon, L.Y.; Singleton, P.A.; Diedrich, F.; Stern, R.; Gilad, E. CD44 interaction with Na+-H+ exchanger (NHE1) creates acidic microenvironments leading to hyaluronidase-2 and cathepsin B activation and breast tumor cell invasion. J. Biol. Chem. 2004, 279, 26991–27007.
- Triantafilou, M.; Miyake, K.; Golenbock, D.T.; Triantafilou, K. Mediators of innate immune recognition of bacteria concentrate in lipid rafts and facilitate lipopolysaccharide-induced cell activation. J. Cell Sci. 2002, 115, 2603–2611.
- Grass, G.D.; Dai, L.; Qin, Z.; Parsons, C.; Toole, B.P. CD147: Regulator of hyaluronan signaling in invasiveness and chemoresistance. Adv. Cancer Res. 2014, 123, 351–373.
- Wu, A.; Wojtowicz, K.; Savary, S.; Hamon, Y.; Trombik, T. Do ABC transporters regulate plasma membrane organization? Cell. Mol. Biol. Lett. 2020, 25, 37.
- Xu, Y.; Yu, Q. E-cadherin negatively regulates CD44-hyaluronan interaction and CD44-mediated tumor invasion and branching morphogenesis. J. Biol. Chem. 2003, 278, 8661–8668.
- Guo, Y.J.; Lin, S.C.; Wang, J.H.; Bigby, M.; Sy, M.S. Palmitoylation of CD44 interferes with CD3-mediated signaling in human T lymphocytes. Int. Immunol. 1994, 6, 213–221.
- Freeman, S.A.; Vega, A.; Riedl, M.; Collins, R.F.; Ostrowski, P.P.; Woods, E.C.; Bertozzi, C.R.; Tammi, M.I.; Lidke, D.S.; Johnson, P.; et al. Transmembrane Pickets Connect Cyto- and Pericellular Skeletons Forming Barriers to Receptor Engagement. Cell 2018, 172, 305–317.e10.
- Hu, S.; Shi, X.; Liu, Y.; He, Y.; Du, Y.; Zhang, G.; Yang, C.; Gao, F. CD44 cross-linking increases malignancy of breast cancer via upregulation of p-Moesin. Cancer Cell Int. 2020, 20, 563.
- Brown, K.L.; Birkenhead, D.; Lai, J.C.; Li, L.; Li, R.; Johnson, P. Regulation of hyaluronan binding by F-actin and colocalization of CD44 and phosphorylated ezrin/radixin/moesin (ERM) proteins in myeloid cells. Exp. Cell Res. 2005, 303, 400–414.
- Liu, D.; Liu, T.; Sy, M.S. Identification of two regions in the cytoplasmic domain of CD44 through which PMA, calcium, and foskolin differentially regulate the binding of CD44 to hyaluronic acid. Cell Immunol. 1998, 190, 132–140.
- Seluanov, A.; Hine, C.; Azpurua, J.; Feigenson, M.; Bozzella, M.; Mao, Z.; Catania, K.C.; Gorbunova, V. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc. Natl. Acad. Sci. USA 2009, 106, 19352–19357.
- Tian, X.; Azpurua, J.; Hine, C.; Vaidya, A.; Myakishev-Rempel, M.; Ablaeva, J.; Mao, Z.; Nevo, E.; Gorbunova, V.; Seluanov, A. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 2013, 499, 346–349.
- Morrison, H.; Sherman, L.S.; Legg, J.; Banine, F.; Isacke, C.; Haipek, C.A.; Gutmann, D.H.; Ponta, H.; Herrlich, P. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 2001, 15, 968–980.
- Jin, H.; Sperka, T.; Herrlich, P.; Morrison, H. Tumorigenic transformation by CPI-17 through inhibition of a merlin phosphatase. Nature 2006, 442, 576–579.
- Hartmann, M.; Parra, L.M.; Ruschel, A.; Bohme, S.; Li, Y.; Morrison, H.; Herrlich, A.; Herrlich, P. Tumor Suppressor NF2 Blocks Cellular Migration by Inhibiting Ectodomain Cleavage of CD44. Mol. Cancer Res. 2015, 13, 879–890.
- Ooki, T.; Murata-Kamiya, N.; Takahashi-Kanemitsu, A.; Wu, W.; Hatakeyama, M. High-Molecular-Weight Hyaluronan Is a Hippo Pathway Ligand Directing Cell Density-Dependent Growth Inhibition via PAR1b. Dev. Cell 2019, 49, 590–604.e9.
- Lichtenthaler, S.F.; Lemberg, M.K.; Fluhrer, R. Proteolytic ectodomain shedding of membrane proteins in mammals-hardware, concepts, and recent developments. EMBO J. 2018, 37, e99456.
- Hartmann, M.; Parra, L.M.; Ruschel, A.; Lindner, C.; Morrison, H.; Herrlich, A.; Herrlich, P. Inside-out Regulation of Ectodomain Cleavage of Cluster-of-Differentiation-44 (CD44) and of Neuregulin-1 Requires Substrate Dimerization. J. Biol. Chem. 2015, 290, 17041–17054.
- Wöhner, B.; Li, W.; Hey, S.; Drobny, A.; Werny, L.; Becker-Pauly, C.; Lucius, R.; Zunke, F.; Linder, S.; Arnold, P. Proteolysis of CD44 at the cell surface controls a downstream protease network. Front. Mol. Biosci. 2023, 10, 1026810.
- Suenaga, N.; Mori, H.; Itoh, Y.; Seiki, M. CD44 binding through the hemopexin-like domain is critical for its shedding by membrane-type 1 matrix metalloproteinase. Oncogene 2005, 24, 859–868.
- Terawaki, S.; Kitano, K.; Aoyama, M.; Mori, T.; Hakoshima, T. MT1-MMP recognition by ERM proteins and its implication in CD44 shedding. Genes Cells 2015, 20, 847–859.
- Tamada, M.; Nagano, O.; Tateyama, S.; Ohmura, M.; Yae, T.; Ishimoto, T.; Sugihara, E.; Onishi, N.; Yamamoto, T.; Yanagawa, H.; et al. Modulation of glucose metabolism by CD44 contributes to antioxidant status and drug resistance in cancer cells. Cancer Res. 2012, 72, 1438–1448.
- Christofk, H.R.; Vander Heiden, M.G.; Harris, M.H.; Ramanathan, A.; Gerszten, R.E.; Wei, R.; Fleming, M.D.; Schreiber, S.L.; Cantley, L.C. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 2008, 452, 230–233.
- Christofk, H.R.; Vander Heiden, M.G.; Wu, N.; Asara, J.M.; Cantley, L.C. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 2008, 452, 181–186.
- Hitosugi, T.; Kang, S.; Vander Heiden, M.G.; Chung, T.W.; Elf, S.; Lythgoe, K.; Dong, S.; Lonial, S.; Wang, X.; Chen, G.Z.; et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci. Signal. 2009, 2, ra73.
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033.
- Matoba, S.; Kang, J.G.; Patino, W.D.; Wragg, A.; Boehm, M.; Gavrilova, O.; Hurley, P.J.; Bunz, F.; Hwang, P.M. p53 regulates mitochondrial respiration. Science 2006, 312, 1650–1653.
- Miletti-Gonzalez, K.E.; Murphy, K.; Kumaran, M.N.; Ravindranath, A.K.; Wernyj, R.P.; Kaur, S.; Miles, G.D.; Lim, E.; Chan, R.; Chekmareva, M.; et al. Identification of function for CD44 intracytoplasmic domain (CD44-ICD): Modulation of matrix metalloproteinase 9 (MMP-9) transcription via novel promoter response element. J. Biol. Chem. 2012, 287, 18995–19007.
- Gao, R.; Li, D.; Xun, J.; Zhou, W.; Li, J.; Wang, J.; Liu, C.; Li, X.; Shen, W.; Qiao, H.; et al. CD44ICD promotes breast cancer stemness via PFKFB4-mediated glucose metabolism. Theranostics 2018, 8, 6248–6262.
