Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Shangbang Gao | -- | 2227 | 2024-01-10 07:13:41 | | | |
| 2 | Jessie Wu | + 3 word(s) | 2230 | 2024-01-10 07:35:27 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Chen, L.; Zhang, S.; Liu, S.; Gao, S. Caenorhabditis elegans Models in Amyotrophic Lateral Sclerosis Mechanism. Encyclopedia. Available online: https://encyclopedia.pub/entry/53640 (accessed on 07 February 2026).
Chen L, Zhang S, Liu S, Gao S. Caenorhabditis elegans Models in Amyotrophic Lateral Sclerosis Mechanism. Encyclopedia. Available at: https://encyclopedia.pub/entry/53640. Accessed February 07, 2026.
Chen, Lili, Shumei Zhang, Sai Liu, Shangbang Gao. "Caenorhabditis elegans Models in Amyotrophic Lateral Sclerosis Mechanism" Encyclopedia, https://encyclopedia.pub/entry/53640 (accessed February 07, 2026).
Chen, L., Zhang, S., Liu, S., & Gao, S. (2024, January 10). Caenorhabditis elegans Models in Amyotrophic Lateral Sclerosis Mechanism. In Encyclopedia. https://encyclopedia.pub/entry/53640
Chen, Lili, et al. "Caenorhabditis elegans Models in Amyotrophic Lateral Sclerosis Mechanism." Encyclopedia. Web. 10 January, 2024.
Copy Citation
Amyotrophic Lateral Sclerosis (ALS) is a debilitating neurodegenerative condition characterized by the progressive degeneration of motor neurons. Despite extensive research in various model animals, the cellular signal mechanisms of ALS remain elusive, impeding the development of efficacious treatments. Among these models, a well-characterized and diminutive organism, Caenorhabditis elegans (C. elegans), has emerged as a potent tool for investigating the molecular and cellular dimensions of ALS pathogenesis.
ALS
cellular mechanism
therapeutic application
1. Brief Introduction of Amyotrophic Lateral Sclerosis
Motor Neuron Disease (MND) constitutes a group of disorders, including, but not limited to, Amyotrophic Lateral Sclerosis (ALS), Progressive Spinal Muscular Atrophy, Primary Lateral Sclerosis, and Progressive Bulbar Palsy. These disorders share a common feature: damage to upper and lower motor neurons, resulting in the loss of essential motor function [1][2]. Typically, individuals diagnosed with MND manifest symptoms such as muscle wasting and limb weakness. In addition to motor impairments, they may also encounter challenges related to language and swallowing [3]. As MND progresses, most patients succumb to complications such as pneumonia or respiratory failure [4]. This research specifically addresses the intricate mechanisms underlying ALS, recognized as the most prevalent adult-onset neurodegenerative form of MND. Researchers predominantly summarize the molecular and cellular pathways from studies conducted with the small model animal Caenorhabditis elegans (C. elegans).
ALS, commonly known as Charcot’s disease or Lou Gehrig’s disease, is a motor neuron disease (MND) affecting both upper and lower motor neurons within the central nervous system, governing voluntary muscle movement. Clinically, ALS is characterized by muscle rigidity and the gradual weakening of limbs and bulbar muscles, leading to varying degrees of difficulty in speech, swallowing, and respiration [4][5]. It is noteworthy that functions such as bladder control, bowel movements, and eye movements typically remain unaffected until the advanced stages of the disease [6]. In addition to muscle dysfunction, 30–50% of ALS patients present with cognitive and other nervous system deficits. Common cognitive symptoms in ALS patients include challenges in social cognition, verbal memory, language, and executive function [7]. Approximately 15% of cases with observed cognitive impairment exhibit visible atrophy in the frontal and/or temporal lobes, resulting in behavioral changes or language impairment meeting the criteria for frontotemporal dementia (FTD) [8]. These observations indicate the complexity of ALS as a progressive disease involving functional deficits in multiple tissues.
2. Pathogenic Mechanisms of Amyotrophic Lateral Sclerosis Implicated in Caenorhabditis elegans
- (1)
-
Innate immunity
Data from clinical studies show that multiple genetic mutations linked to ALS enhance neuroinflammation, which provides compelling evidence for immune dysregulation in the pathogenesis of ALS [9][10][11]. Although C. elegans lacks a classical inflammatory response or inflammatory cytokines analogous to mammals, it possesses an innate immune system. Notably, mutated ALS-associated proteins have been found to activate an innate immune response in C. elegans [12][13][14]. In C. elegans strains expressing mutant TDP-43 or FUS in their motor neurons, age-dependent motility defects culminate in paralysis and motor neuron degeneration at a rate significantly higher than that observed in wild-type TDP-43 or FUS control strains. By examining the expression of immune response proteins, including NLP-29 (an antimicrobial, neuropeptide-like protein expressed in hypodermal and intestinal tissue), it is evident that the expression of mutant TDP-43A315T or FUSS57∆ protein triggers the upregulation of immune response genes, suggesting that innate immune response may contribute to motor neuron neurodegeneration. Furthermore, mutated ALS-associated proteins trigger an TIR-1/Sarm1 immune pathway innate immune response in C. elegans motor neurons [15][16]. Loss-of-function mutations in tir-1, associated downstream kinases, and the transcription factor atf-7 collectively serve to suppress motor neuron degeneration, further supporting the notion that the innate immune system is involved in ALS models.
Despite knowing the importance of the immune response in ALS, there are many details that have not yet been elucidated. For instance, mutated ALS-associated proteins in neurons may elicit an immune response as part of a host defense reaction against pathogens or aid tissue repair. The molecules necessary to induce the expression of NLP-29 also need to be further explored. Chikka et al. reported that activation of the mitochondrial p38MAPK/ATF-7 immune pathway in the intestine is neuroprotective and sufficient to prevent rotenone-induced neurodegeneration [17]. Due to mitochondrial dysfunction being a prevalent feature of many neurodegenerative diseases, including ALS [18], the mitochondria-regulated immune pathway may also be involved in C. elegans motor neuron degeneration. C. elegans’ innate immune response coordinates its activity with the insulin/IGF-1 pathway [19], suggesting that insulin-related immune pathways are also worth investigating. Nevertheless, these studies reveal that cell-based strategies that enhance anti-inflammatory reactivity and reverse immune dysregulation offer the potential to slow disease progression and improve the quality of life of patients with ALS.
- (2)
-
Autophagy
It is widely acknowledged that the dysregulation of autophagy in motor neurons is a pivotal event in ALS [20][21][22]. Particularly, intensified immunoreactivity in the cytoplasm of motor neurons for microtubule-associated protein 1 light chain 3 (LC3), which is a marker of autophagosome, is frequently observed in the spinal motor neurons of ALS patients [23][24]. Consistent with this observation, C. elegans with dynactin 1 knockdown (dnc-1(RNAi) worms) in ventral motor neurons under the control of the Pacr-2 promoter exhibited notable motor impairments, coupled with axonal and neuronal degeneration. Notably, the autophagosomes were easily trapped where the axon was tight, curved, or at spheroids. The phenomenon was followed by the accumulation of autophagosomes distal to the trapped sites [25]. Given that autophagosomes serve as cargo for dynein/dynactin complexes and play a pivotal role in the turnover of various organelles and proteins, the accumulation of autophagosomes suggests a potential contribution of dysfunctional autophagy to motor neuron degeneration in ALS. Indeed, the introduction of pharmacological disruptions to autophagy, using 3-MA, resulted in locomotory defects and axonal degeneration mirroring those observed in dnc-1(RNAi) worms. This implies that a compromised autophagy system alone is adequate to induce motor neuronal degeneration [25].
The contribution of defective autophagy to neuronal dysfunction in ALS is well-documented by autophagy-related genes [9][26][27][28][29][30]. The selectivity of autophagy is mediated by autophagy receptors that recognize and deliver cargoes to autophagosomes for degradation. SQSTM1/p62 is an autophagy receptor that is commonly found in protein aggregates in ALS brains. Related reports showed that SQSTM1 promotes the clearance of stress granules, a hallmark of ALS, via selective autophagy [31][32]. The main autophagy process is proteotoxic stress, which activates serine/threonine kinase TBK1, promotes phosphorylation of autophagy receptor SQSTM1, and activates selective autophagy. In contrast, ALS-linked mutations of TBK1 or SQSTM1 reduce SQSTM1 phosphorylation and compromise ubiquitinated cargo binding and clearance (Figure 1). The accumulation of SQSTM1 implicates a disturbance of the selective autophagy pathway [33]. Corresponding with the accumulation of autophagosomes, SQSTM1/p62 has been observed to accumulate in the motor neurons of ALS patients [34]. This observation aligns with findings demonstrating elevated levels of LC3-positive autophagy vesicles in the motor neurons of ALS patients with FUS mutations [35]. Notably, in an ALS C. elegans model involving overexpressing human mutant FUS proteins, a gain of toxic function mechanism disrupts basal neuronal autophagy. There was an increased accumulation of SQST-1 that disrupts neuromuscular function in stress conditions, the C. elegans ortholog for SQSTM1/p62, in motor neurons [36], reinforcing the link between autophagy dysfunction and ALS pathology. Conversely, the loss of sqst-1 suppresses both neuromuscular and stress-induced locomotion defects in FUS-associated ALS worms. It is worth mentioning that this suppression likely does not accompany a correction of neuronal autophagy defects [36], indicating that SQST-1 operates through an autophagy-independent pathway or alternative mechanisms to ameliorate ALS-related locomotion impairments [37][38]. The mutation of a single autophagy receptor can induce the decline of autophagy and lead to abnormal protein accumulation. But when autophagy receptors are passively increased, reducing autophagy levels may have a positive effect. Thus, the treatment of ALS requires multi-factorial and systematic consideration.
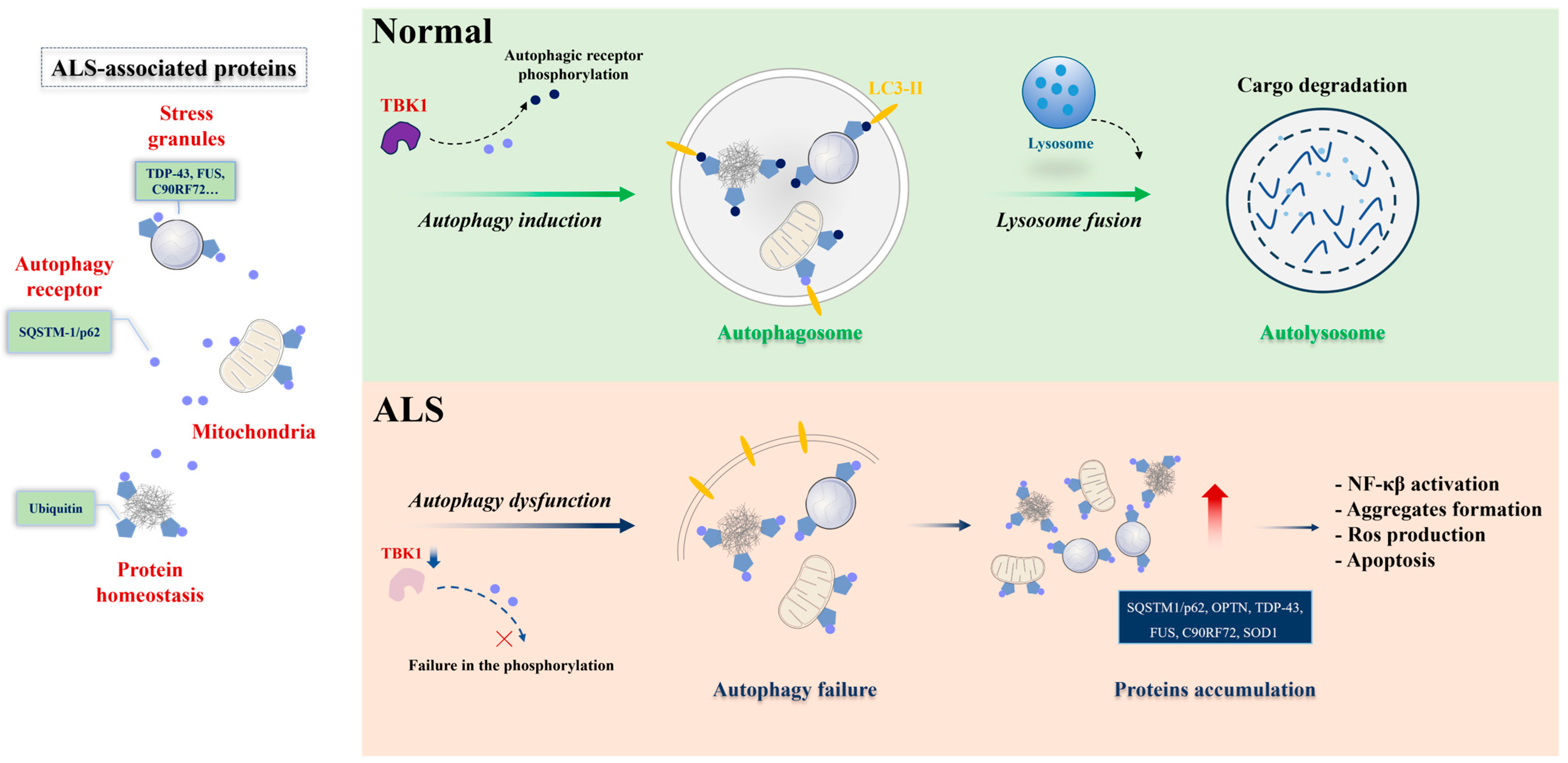
Figure 1. Selective autophagy under physiological and ALS pathological conditions. Protein aggregates, stress granules, and dysfunctional mitochondria serve as substrates for selective autophagy degradation. In physiological conditions (upper panel), these substrates are bound by selective autophagy receptors, such as SQSTM-1/p62 (represented in blue circle), via ubiquitin-binding domains (ubiquitin, in pentagon). The selective autophagy receptors associate with LC3-II proteins in the autophagosome (represented in yellow) or other members of the autophagy machinery. Posttranslational modifications in the receptors can enhance binding with ubiquitinated substrates or the LC3-II protein. TBK1 is one of the main kinases acting in this process. The cargo-receptor- LC3-II complexes are then sequestered by de novo double-membrane vesicles called the autophagosome, which fuses with the lysosome for the final degradation. Under ALS conditions (lower panel), failure in selective autophagy can occur through mutations in the genes encoding the receptors themselves or in the kinase, reducing the activity of the pathway and promoting the accumulation of toxic substrates for motor neurons. Figure was generated by PowerPoint 2013.
Furthermore, increased expression levels of autophagic genes by daf-2(e1370) have been shown to protect C. elegans motor neurons against the toxicity of human SOD-1(G93A) [39]. Metformin, the globally prescribed biguanide drug worldwide for the treatment of type II diabetes, alleviates motor dysfunction in human SOD-1(G93A)-associated ALS worms, partly through enhancement of autophagy [40]. Although not explicitly validated in C. elegans models, investigations in other model systems have demonstrated cross-regulation between TDP-43 pathology and autophagy [41][42]. These findings imply the existence of supplementary autophagy mechanisms in ALS [43]. In conclusion, enhancing autophagy emerges as a novel and significant therapeutic target for addressing motor neuron degeneration in ALS.
- (3)
-
Protein homeostasis
Protein homeostasis (proteostasis) is carefully maintained through a finely regulated and interconnected network of biological pathways, crucial for preventing the accumulation and aggregation of damaged or misfolded proteins [44]. Conversely, the breakdown of proteostasis has been implicated in the etiology of various neurodegenerative diseases, including ALS [45]. Zhang et al. conducted genetic analysis and expression profiling of loss-of-function tdp-1 mutants, elucidating the role of C. elegans TDP-1 (nematode TDP-43 ortholog) in regulating protein homeostasis. In diverse proteotoxicity models, the loss of TDP-1 alleviated protein aggregation and neuronal dysfunction. Their findings suggest that TDP-1 loss may modify global RNA levels, consequently impacting protein homeostasis and prompting cellular adaptation to stress on protein quality control systems [46]. Transgenic C. elegans models that express human TDP-43 variants displayed severe locomotor defects associated with the aggregation of TDP-43 in neurons [47]. Notably, the neurotoxicity and protein aggregation of TDP-43 were influenced by environmental temperature, and heat shock transcriptional factor 1 (HSF-1) played a role through protein quality control [48], indicating that a deficiency in protein quality control serves as a risk factor for TDP-43-associated ALS [47].
Aging-related neurodegeneration associated with TDP-43 is further connected to protein misfolding. The well-explored regulatory mechanism governing longevity and proteostasis involves the modulation of the insulin/IGF-1 signaling pathway through phosphorylation [49][50]. In the nematode C. elegans, the downstream receptor of insulin molecule, daf-2, has demonstrated the capability to counteract the shortened lifespan resulting from FUS overexpression [47][51]. These findings illuminate the intricate network of cellular mechanisms, notably the insulin/IGF-1 pathway, designed to preserve protein homeostasis in the presence of environmentally induced damage or genetically encoded misfolded proteins.
- (4)
-
Energy metabolism
While the intricate mechanisms underlying the association between mutant TDP-43, FUS, and Amyotrophic Lateral Sclerosis (ALS) are intricate and multifaceted, an accumulating body of evidence supports the presence of dysregulated energy metabolism in both ALS patients and relevant models [52]. The AMP-activated protein kinase (AMPK) serves as a pivotal cellular energy sensor. Upon activation, AMPK restores energy homeostasis by facilitating catabolic pathways, thereby promoting ATP generation [53]. Notably, heightened AMPK activation has been documented in the motor neurons of ALS patients, displaying a notable correlation with the extent of cytoplasmic mislocalization of TDP-43 [54]. These observations establish a clear link between energy depletion in human motor neurons and the pathological presence of TDP-43 in ALS. In line with this correlation, reducing AMPK activity has been demonstrated to ameliorate disease outcomes both in vitro and in C. elegans models expressing mutant SOD1 or TDP-43 [55]. Although a definitive mechanistic link between AMPK-regulated energy metabolism and TDP-43 mislocalization remains elusive, a proposed hypothesis suggests that the aggregation of TDP-43 stems from AMPK-mediated inhibition of nucleocytoplasmic transport.
Mitochondrial dysfunction is a prevalent characteristic of ALS [56][57]. Mutant forms of TDP-43, SOD-1, and FUS proteins have been implicated in disrupting mitochondrial structure and function [58][59][60]. This dysfunction induces an energy imbalance within neurons, affecting energy production and utilization. Specifically, FUS mutations have been linked to disturbances in mitochondrial function, potentially impeding the neurons’ capacity to generate ATP [58]. Consequently, compromised energy metabolism may play a role in the vulnerability and degeneration of motor neurons in ALS. Enhancing mitochondrial biogenesis emerges as an appealing therapeutic strategy for ALS. It is important to note that, despite the absence of current evidence from the C. elegans model, gaining further insights into these altered physiological processes in neurons—particularly by expressing mutant TDP-43 or FUS in C. elegans—becomes crucial for a more comprehensive understanding of ALS pathogenesis, specifically pertaining to energy metabolism.
In summary, mitochondria play a key role in ATP supply to cells via oxidative phosphorylation. Decreased ATP levels emerge as a common feature in ALS. It is conceivable that, in line with the high energy demands of neurons, gradual depletion of ATP, due to reduced respiration, may trigger neuronal degeneration [18][52]. In addition, the mitochondrial REDOX reaction is associated with the production of SOD. A lower concentration of ROS is essential for normal cellular signaling, whereas a higher concentration and long-term exposure of ROS cause damage to cellular macromolecules such as DNA, lipids, and proteins, ultimately resulting in necrosis and apoptotic cell death [61]. Altogether, these data suggest that bioenergetic abnormalities are likely to be pathophysiologically relevant to ALS disease.
References
- Arora, R.D.; Khan, Y.S. Motor Neuron Disease. In StatPearls; Disclosure: Yusuf Khan Declares No Relevant Financial Relationships with Ineligible Companies; Ineligible Companies: Treasure Island, FL, USA, 2023.
- Dugger, B.N.; Dickson, D.W. Pathology of Neurodegenerative Diseases. Cold Spring Harb. Perspect. Biol. 2017, 9, a028035.
- Chen, S.; Sayana, P.; Zhang, X.; Le, W. Genetics of amyotrophic lateral sclerosis: An update. Mol. Neurodegener. 2013, 8, 28.
- Wijesekera, L.C.; Leigh, P.N. Amyotrophic lateral sclerosis. Orphanet J. Rare Dis. 2009, 4, 3.
- Zarei, S.; Carr, K.; Reiley, L.; Diaz, K.; Guerra, O.; Altamirano, P.F.; Pagani, W.; Lodin, D.; Orozco, G.; Chinea, A. A comprehensive review of amyotrophic lateral sclerosis. Surg. Neurol. Int. 2015, 6, 171.
- Hobson, E.V.; McDermott, C.J. Supportive and symptomatic management of amyotrophic lateral sclerosis. Nat. Rev. Neurol. 2016, 12, 526–538.
- Beeldman, E.; Raaphorst, J.; Klein Twennaar, M.; de Visser, M.; Schmand, B.A.; de Haan, R.J. The cognitive profile of ALS: A systematic review and meta-analysis update. J. Neurol. Neurosurg. Psychiatry 2016, 87, 611–619.
- Ringholz, G.M.; Appel, S.H.; Bradshaw, M.; Cooke, N.A.; Mosnik, D.M.; Schulz, P.E. Prevalence and patterns of cognitive impairment in sporadic ALS. Neurology 2005, 65, 586–590.
- Maruyama, H.; Morino, H.; Ito, H.; Izumi, Y.; Kato, H.; Watanabe, Y.; Kinoshita, Y.; Kamada, M.; Nodera, H.; Suzuki, H.; et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature 2010, 465, 223–226.
- McCombe, P.A.; Henderson, R.D. The Role of Immune and Inflammatory Mechanisms in ALS. Curr. Mol. Med. 2011, 11, 246–254.
- Ravits, J.; Appel, S.; Baloh, R.H.; Barohn, R.; Brooks, B.R.; Elman, L.; Floeter, M.K.; Henderson, C.; Lomen-Hoerth, C.; Macklis, J.D.; et al. Deciphering amyotrophic lateral sclerosis: What phenotype, neuropathology and genetics are telling us about pathogenesis. Amyotroph. Lateral Scler. Frontotemporal. Degener. 2013, 14 (Suppl. 1), 5–18.
- Veriepe, J.; Fossouo, L.; Parker, J.A. Neurodegeneration in C. elegans models of ALS requires TIR-1/Sarm1 immune pathway activation in neurons. Nat. Commun. 2015, 6, 7319.
- Beers, D.R.; Appel, S.H. Immune dysregulation in amyotrophic lateral sclerosis: Mechanisms and emerging therapies. Lancet Neurol. 2019, 18, 211–220.
- Heneka, M.T.; Kummer, M.P.; Latz, E. Innate immune activation in neurodegenerative disease. Nat. Rev. Immunol. 2014, 14, 463–477.
- Pujol, N.; Zugasti, O.; Wong, D.; Couillault, C.; Kurz, C.L.; Schulenburg, H.; Ewbank, J.J. Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathog. 2008, 4, e1000105.
- Couillault, C.; Pujol, N.; Reboul, J.; Sabatier, L.; Guichou, J.F.; Kohara, Y.; Ewbank, J.J. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 2004, 5, 488–494.
- Chikka, M.R.; Anbalagan, C.; Dvorak, K.; Dombeck, K.; Prahlad, V. The Mitochondria-Regulated Immune Pathway Activated in the Intestine Is Neuroprotective. Cell Rep. 2016, 16, 2399–2414.
- Smith, E.F.; Shaw, P.J.; De Vos, K.J. The role of mitochondria in amyotrophic lateral sclerosis. Neurosci. Let. 2019, 710, 132933.
- Singh, V.; Aballay, A. Regulation of DAF-16-mediated Innate Immunity in. J. Biol. Chem. 2009, 284, 35580–35587.
- Ravikumar, B.; Acevedo-Arozena, A.; Imarisio, S.; Berger, Z.; Vacher, C.; O’Kane, C.J.; Brown, S.D.; Rubinsztein, D.C. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat. Genet. 2005, 37, 771–776.
- Komatsu, M.; Wang, Q.J.; Holstein, G.R.; Friedrich, V.L., Jr.; Iwata, J.; Kominami, E.; Chait, B.T.; Tanaka, K.; Yue, Z. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc. Natl. Acad. Sci. USA 2007, 104, 14489–14494.
- Nguyen, H.Q.; Zada, S.; Lai, T.H.; Pham, T.M.; Hwang, J.S.; Ahmed, M.; Kim, D.R. Calpain-dependent Beclin1 cleavage stimulates senescence-associated cell death in HT22 hippocampal cells under the oxidative stress conditions. Neurosci. Lett. 2019, 701, 106–111.
- Sasaki, S. Autophagy in spinal cord motor neurons in sporadic amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 2011, 70, 349–359.
- Li, L.; Zhang, X.; Le, W. Altered macroautophagy in the spinal cord of SOD1 mutant mice. Autophagy 2008, 4, 290–293.
- Ikenaka, K.; Kawai, K.; Katsuno, M.; Huang, Z.; Jiang, Y.M.; Iguchi, Y.; Kobayashi, K.; Kimata, T.; Waza, M.; Tanaka, F.; et al. dnc-1/dynactin 1 knockdown disrupts transport of autophagosomes and induces motor neuron degeneration. PLoS ONE 2013, 8, e54511.
- Fecto, F.; Yan, J.; Vemula, S.P.; Liu, E.; Yang, Y.; Chen, W.; Zheng, J.G.; Shi, Y.; Siddique, N.; Arrat, H.; et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch. Neurol. 2011, 68, 1440–1446.
- Deng, H.X.; Chen, W.; Hong, S.T.; Boycott, K.M.; Gorrie, G.H.; Siddique, N.; Yang, Y.; Fecto, F.; Shi, Y.; Zhai, H.; et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 2011, 477, 211–215.
- Buchan, J.R.; Kolaitis, R.M.; Taylor, J.P.; Parker, R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 2013, 153, 1461–1474.
- Wong, Y.C.; Holzbaur, E.L. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl. Acad. Sci. USA 2014, 111, E4439–E4448.
- Freischmidt, A.; Wieland, T.; Richter, B.; Ruf, W.; Schaeffer, V.; Muller, K.; Marroquin, N.; Nordin, F.; Hubers, A.; Weydt, P.; et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat. Neurosci. 2015, 18, 631–636.
- Guo, H.S.; Chitiprolu, M.; Gagnon, D.; Meng, L.R.; Perez-Iratxeta, C.; Lagace, D.; Gibbings, D. Autophagy supports genomic stability by degrading retrotransposon RNA. Nat. Commun. 2014, 5, 5276.
- Chitiprolu, M.; Jagow, C.; Tremblay, V.; Bondy-Chorney, E.; Paris, G.; Savard, A.; Palidwor, G.; Barry, F.A.; Zinman, L.; Keith, J.; et al. A complex of C9ORF72 and p62 uses arginine methylation to eliminate stress granules by autophagy. Nat. Commun. 2018, 9, 2794.
- Deng, Z.Q.; Lim, J.; Wang, Q.; Purtell, K.; Wu, S.; Palomo, G.M.; Tan, H.Y.; Manfredi, G.; Zhao, Y.X.; Peng, J.M.; et al. ALS-FTLD-linked mutations of SQSTM1/p62 disrupt selective autophagy and NFE2L2/NRF2 anti-oxidative stress pathway. Autophagy 2020, 16, 917–931.
- Blokhuis, A.M.; Groen, E.J.; Koppers, M.; van den Berg, L.H.; Pasterkamp, R.J. Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol. 2013, 125, 777–794.
- Soo, K.Y.; Sultana, J.; King, A.E.; Atkinson, R.; Warraich, S.T.; Sundaramoorthy, V.; Blair, I.; Farg, M.A.; Atkin, J.D. ALS-associated mutant FUS inhibits macroautophagy which is restored by overexpression of Rab1. Cell Death Discov. 2015, 1, 15030.
- Baskoylu, S.N.; Chapkis, N.; Unsal, B.; Lins, J.; Schuch, K.; Simon, J.; Hart, A.C. Disrupted autophagy and neuronal dysfunction in C. elegans knockin models of FUS amyotrophic lateral sclerosis. Cell Rep. 2022, 38, 110195.
- Gomez-Virgilio, L.; Silva-Lucero, M.D.; Flores-Morelos, D.S.; Gallardo-Nieto, J.; Lopez-Toledo, G.; Abarca-Fernandez, A.M.; Zacapala-Gomez, A.E.; Luna-Munoz, J.; Montiel-Sosa, F.; Soto-Rojas, L.O.; et al. Autophagy: A Key Regulator of Homeostasis and Disease: An Overview of Molecular Mechanisms and Modulators. Cells 2022, 11, 2262.
- Chua, J.P.; De Calbiac, H.; Kabashi, E.; Barmada, S.J. Autophagy and ALS: Mechanistic insights and therapeutic implications. Autophagy 2022, 18, 254–282.
- Li, J.; Huang, K.X.; Le, W.D. Establishing a novel C. elegans model to investigate the role of autophagy in amyotrophic lateral sclerosis. Acta Pharmacol. Sin. 2013, 34, 644–650.
- Xu, H.; Jia, C.C.; Cheng, C.; Wu, H.F.; Cai, H.B.; Le, W.D. Activation of autophagy attenuates motor deficits and extends lifespan in a model of ALS. Free Radic. Biol. Med. 2022, 181, 52–61.
- Barmada, S.J.; Serio, A.; Arjun, A.; Bilican, B.; Daub, A.; Ando, D.M.; Tsvetkov, A.; Pleiss, M.; Li, X.L.; Peisach, D.; et al. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat. Chem. Biol. 2014, 10, 677–685.
- Xia, Q.; Wang, H.F.; Hao, Z.B.; Fu, C.; Hu, Q.S.; Gao, F.; Ren, H.G.; Chen, D.; Han, J.H.; Ying, Z.; et al. TDP-43 loss of function increases TFEB activity and blocks autophagosome-lysosome fusion. EMBO J. 2016, 35, 121–142.
- Wong, S.Q.; Kumar, A.V.; Mills, J.; Lapierre, L.R. C. elegans to model autophagy-related human disorders. Prog. Mol. Biol. Transl. Sci. 2020, 172, 325–373.
- Noormohammadi, A.; Calculli, G.; Gutierrez-Garcia, R.; Khodakarami, A.; Koyuncu, S.; Vilchez, D. Mechanisms of protein homeostasis (proteostasis) maintain stem cell identity in mammalian pluripotent stem cells. Cell. Mol. Life Sci. 2018, 75, 275–290.
- Hohn, A.; Tramutola, A.; Cascella, R. Proteostasis Failure in Neurodegenerative Diseases: Focus on Oxidative Stress. Oxid. Med. Cell. Longev. 2020, 2020, 5497046.
- Zhang, T.; Hwang, H.Y.; Hao, H.; Talbot, C., Jr.; Wang, J. Caenorhabditis elegans RNA-processing protein TDP-1 regulates protein homeostasis and life span. J. Biol. Chem. 2012, 287, 8371–8382.
- Zhang, T.; Mullane, P.C.; Periz, G.; Wang, J. TDP-43 neurotoxicity and protein aggregation modulated by heat shock factor and insulin/IGF-1 signaling. Hum. Mol. Genet. 2011, 20, 1952–1965.
- Kmiecik, S.W.; Mayer, M.P. Molecular mechanisms of heat shock factor 1 regulation. Trends Biochem. Sci. 2022, 47, 218–234.
- Halaschek-Wiener, J.; Khattra, J.S.; McKay, S.; Pouzyrev, A.; Stott, J.M.; Yang, G.S.; Holt, R.A.; Jones, S.J.; Marra, M.A.; Brooks-Wilson, A.R.; et al. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res. 2005, 15, 603–615.
- Chistyakova, O.V.; Bondareva, V.M.; Shipilov, V.N.; Sukhov, I.B.; Shpakov, A.O. Intranasal administration of insulin eliminates the deficit of long-term spatial memory in rats with neonatal diabetes mellitus. Dokl. Biochem. Biophys. 2011, 440, 216–218.
- Boccitto, M.; Lamitina, T.; Kalb, R.G. Daf-2 signaling modifies mutant SOD1 toxicity in C. elegans. PLoS ONE 2012, 7, e33494.
- Vandoorne, T.; De Bock, K.; Van Den Bosch, L. Energy metabolism in ALS: An underappreciated opportunity? Acta Neuropathol. 2018, 135, 489–509.
- Hardie, D.G.; Schaffer, B.E.; Brunet, A. AMPK: An Energy-Sensing Pathway with Multiple Inputs and Outputs. Trends Cell Biol. 2016, 26, 190–201.
- Liu, Y.J.; Ju, T.C.; Chen, H.M.; Jang, Y.S.; Lee, L.M.; Lai, H.L.; Tai, H.C.; Fang, J.M.; Lin, Y.L.; Tu, P.H.; et al. Activation of AMP-activated protein kinase α1 mediates mislocalization of TDP-43 in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2015, 24, 787–801.
- Lim, M.A.; Selak, M.A.; Xiang, Z.; Krainc, D.; Neve, R.L.; Kraemer, B.C.; Watts, J.L.; Kalb, R.G. Reduced Activity of AMP-Activated Protein Kinase Protects against Genetic Models of Motor Neuron Disease. J. Neurosci. 2012, 32, 1123–1141.
- Dupuis, L.; de Aguilar, J.L.G.; Oudart, H.; de Tapia, M.; Barbeito, L.; Loeffler, J.P. Mitochondria in Amyotrophic Lateral Sclerosis: A Trigger and a Target. Neurodegener. Dis. 2004, 1, 245–254.
- Zhao, J.T.; Wang, X.M.; Huo, Z.J.; Chen, Y.C.; Liu, J.M.; Zhao, Z.H.; Meng, F.D.; Su, Q.; Bao, W.W.; Zhang, L.Y.; et al. The Impact of Mitochondrial Dysfunction in Amyotrophic Lateral Sclerosis. Cells 2022, 11, 2049.
- So, E.; Mitchell, J.C.; Memmi, C.; Chennell, G.; Vizcay-Barrena, G.; Allison, L.; Shaw, C.E.; Vance, C. Mitochondrial abnormalities and disruption of the neuromuscular junction precede the clinical phenotype and motor neuron loss in hFUSWT transgenic mice. Hum. Mol. Genet. 2018, 27, 463–474.
- Kong, J.M.; Xu, Z.S. Massive mitochondrial degeneration in motor neurons triggers the onset of amyotrophic lateral sclerosis in mice expressing a mutant SOD1. J. Neurosci. 1998, 18, 3241–3250.
- Shan, X.; Chiang, P.M.; Price, D.L.; Wong, P.C. Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of transgenic mice. Proc. Natl. Acad. Sci. USA 2010, 107, 16325–16330.
- Guillot, S.J.; Bolborea, M.; Dupuis, L. Dysregulation of energy homeostasis in amyotrophic lateral sclerosis. Curr. Opin. Neurol. 2021, 34, 773–780.
More
Information
Subjects:
Cell Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
515
Revisions:
2 times
(View History)
Update Date:
10 Jan 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




